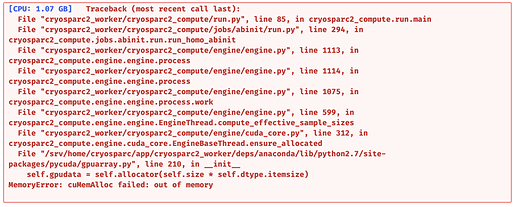
Hi All,
I’ve collected a dataset of a small protein complex (~50 kDa) with surprisingly (to me at least) good contrast, but cannot get them to align in either 2D or 3D.
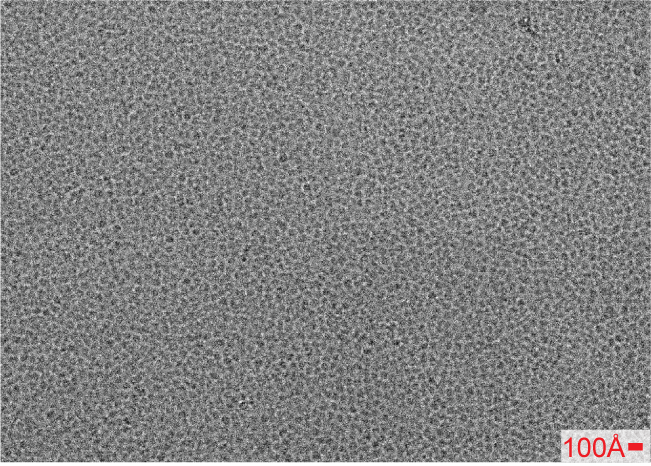
The particles are fundamentally mostly beta propellers that are ~45A in diameter. I’m attaching a 2.5 um defocus micrograph for reference. A bit dense yes, but not too dissimilar to some of the published micrographs that yield high resolution structures.
The images were collected using on a Titan Krios with a K3 and energy filter. The collection strategy was involved using beam-tilt to cover multiple shots per hole on a 3x3 grid, with coma free compensation in SerialEM. Defocus was sampled between 0.8 and 3 um. We used a super-res angpix of 0.326A, and then binned 2x during motion correction. The dose is 60 e/ang^2 over 40 frames. I selected micrographs with ctf fits better than 3.5A and could extract ~12m particles.
The picking was easy with LoG picking, and could be improved with “fuzzy” 2D templates that effectively match the size and shape of the particle (see below). I am convinced that the particles are picked and centered properly.
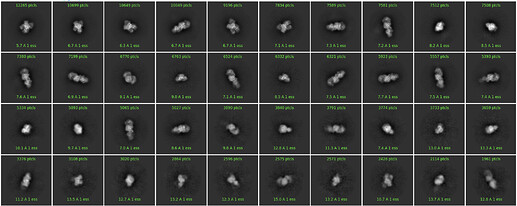
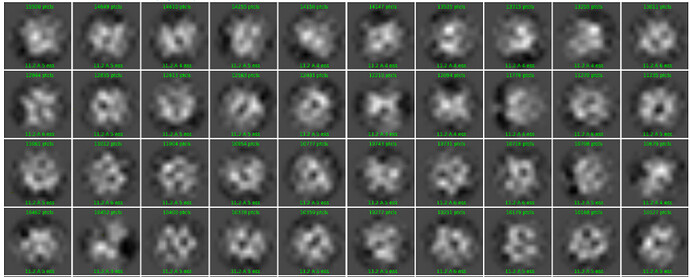
I have tried to run a 2D classification and this is an example subset of classes:
I’ve explored the option parameter space:
- Increased online em cycles to 40
- Increased batch size to 400 or 1000
- Increased class number to 150, 200, 250
- Varied the particle box size between 1.5x and 2x the particle diameter of 45A
- Tightened the inner circular mask to 55A, 60A, 65A
- tried enforcing non-neg and clamp solvent
- Tried limiting the resolution to 8, 10, 12, 15A
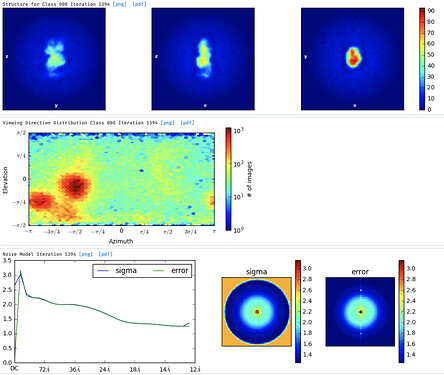
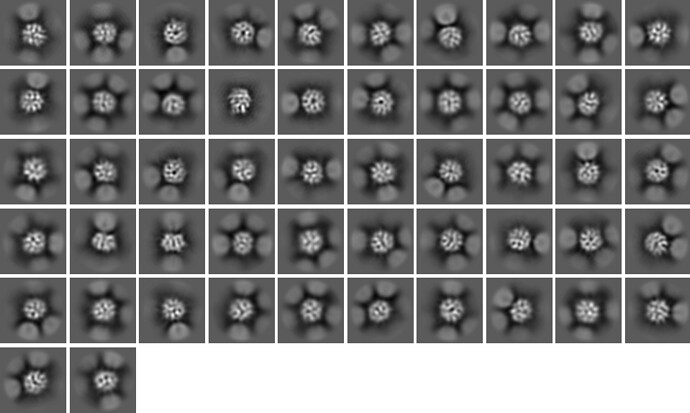
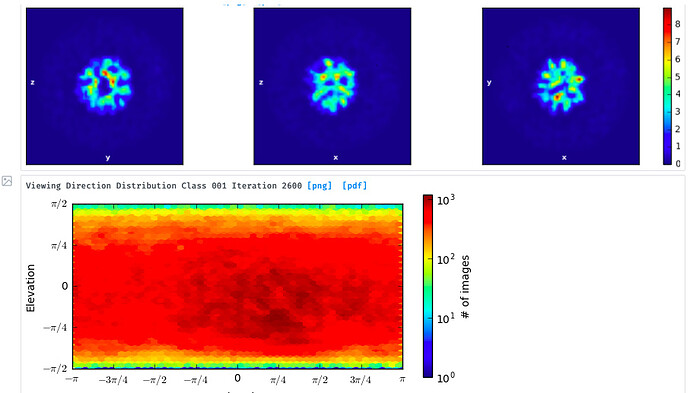
I’ve then tried skipping 2D classification and ab initio reconstruction with either 1, 3 or 6 classes. I tried increasing initial and final minibatch sizes to 1500, as well as limiting the initial resolution to 20A and the final resolution to 9 or 12A. The initial models do not seem right and many show streaky density (overrefined noise?).
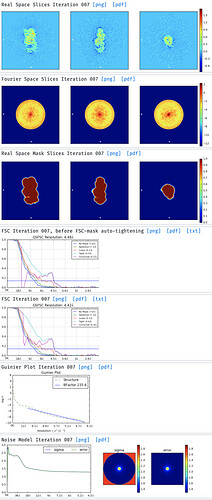
With the particles downsampled to a pixel size of 1.1A, I have also tried homogenous and heterogenous refinement either against these poor inital models, or against a volume calculated from a crystal structure with molmap. I have a crystal structure of the beta propeller core of the particle and am really only interested in the interaction with a peptide that supposedly binds on the surface. I’ve dared go as low as 7A lowpass filtering on the initial model volume, hoping particle alignments might “latch” onto secondary structure features. I’ve tried extracting good particles through heterogenous refinement with one 5/6 initial models having phases randomized to 25A. Nothing to indicate a successful reconstruction or discrimination of good particles.
I’ve tried binning particles to 2.6A pixel size, in an attempt to improve contrast and repeating the some of the refinement steps above, without any success.
I understand that the particles are small and not very feature-rich, but I was excited by the amount of contrast I could see, even at low defocus.
Any ideas or feedback would be helpful and appreciated!
Cheers,
Stefan
 ,
,