Hi, I am getting a similar error when trying to perform a homogeneous refinement job. Any insights on what could be the issue here? Thank you!
Welcome to the forum @himanshumehra11 .
Please can you post additional information:
- Error message as text
- the CryoSPARC version
- how the input volume was prepared. Was the volume created or modified using external software, then imported to CryoSPARC?
Hi, Here’s the additional information:
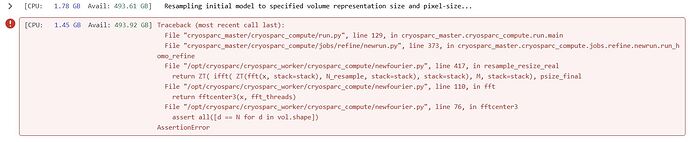
1- Traceback (most recent call last):
File “cryosparc_master/cryosparc_compute/run.py”, line 129, in cryosparc_master.cryosparc_compute.run.main
File “cryosparc_master/cryosparc_compute/jobs/refine/newrun.py”, line 373, in cryosparc_master.cryosparc_compute.jobs.refine.newrun.run_homo_refine
File “/opt/cryosparc/cryosparc_worker/cryosparc_compute/newfourier.py”, line 417, in resample_resize_real
return ZT( ifft( ZT(fft(x, stack=stack), N_resample, stack=stack), stack=stack), M, stack=stack), psize_final
File “/opt/cryosparc/cryosparc_worker/cryosparc_compute/newfourier.py”, line 110, in fft
return fftcenter3(x, fft_threads)
File “/opt/cryosparc/cryosparc_worker/cryosparc_compute/newfourier.py”, line 76, in fftcenter3
assert all([d == N for d in vol.shape])
AssertionError
2- CryoSPARC version v4.7.0
3- Yes, I prepared the input volume using ChimeraX, then imported it as a map.
Is your volume a cube? If not that could be the issue
Thanks @olibclarke. Used your comments from this to fix it: Errors in using an imported volume. Used an ab-initio volume and performed the molmap #1 12 onGrid #2. (#1- PDB model for density map generation, #2- ab initio volume) The density map generated this way doesn’t lead to the error.