Hi, I have imported a mrc volume generated from a pdb file using molmap in Chimera. I got an error below when I did heterogeneous refinement with this volume.
File “cryosparc2_compute/fourier.py”, line 175, in resample_real
fft(x, stack=stack), N, rescale, stack), stack=stack)
File “cryosparc2_compute/fourier.py”, line 110, in fft
return fftcenter3(x, fft_threads)
File “cryosparc2_compute/fourier.py”, line 69, in fftcenter3
assert all([d%2 == 0 for d in vol.shape])
AssertionError
I tried to use Volume Tools on the same imported volume but I got this error this time.
File “cryosparc2_compute/jobs/utilities/run_volume_tools.py”, line 57, in run
assert n.all(n.array(input_shape)==N_input), “Cube volumes only”
AssertionError: Cube volumes only
Any idea what is the problem and how to resolve these errors?
I have another question. When I imported an Ab-initio volume generated from a data set in cryoSPARC2 and used it for refinement for another data set, I got result volumes that did not align to the Ab-initio volume. Is it normal or something is wrong?
Your volume is not a cube. Chimera by default generates molmap mrcs in a minimal box, which can have odd dimensions. You can correct this by resampling on the grid of the original map - e.g. vop resample #map1 ongrid #map2 - or by using molmap with the ongrid option - e.g. molmap sel 20 ongrid #mymap
Cheers
Oli
2 Likes
You definitely want to use molmap ... ongrid. Tom Goddard (lead Chimera developer) found that the interpolation errors were significant otherwise and implemented molmap ... ongrid in response.
1 Like
yes - although I don’t think that matters much with molmap really? I would usually only be using a molmap volume as the starting point for a mask, in which case I’ll usually filter at low res anyway and not care about minor interpolation errors.
Hi guys,
Thank you for the responses. It is working now!
Oli, I found Tom’s mailing list post about the interpolation errors here:
http://plato.cgl.ucsf.edu/pipermail/chimera-users/2016-May/012333.html
2nd para opens with:
I tested this to see how much interpolation (vop resample command) changes the sum of atom Gaussians and I am shocked by the result.
Yes - I know that it does - I just don’t think this matters much, as molmap is only used to make a simulated map from atoms, or am I missing something? The errors introduced by vop resample #map1 ongrid #map2 do matter, because they affect experimentally obtained maps.
Molmap only affects calculated maps, which are only used for mask creation, or maybe in rare cases as an initial model for refinement.
(having said all this - the errors from vop resample are pretty minimal in my experience if using a sufficiently fine grid for the reference map - if the res of all maps is close to the limit from sampling it is a different story)
The person in the chimera-users post was going to use it for map-to-model FSC. Tom found that there was a pretty large effect in using molmap with defaults and then vop resample as opposed to molmap with the right box options, so he added molmap ongrid to make it easier to set the box.
I think you’re right about the use of molmap in general though. In particular it should never be used for map-to-model FSCs! You probably know, but for any one else that happens by the right way to simulate a map is with phenix.fmodel.
# Example with boxsize 256 @ 1.02 A/px.
sed -i '/^CRYST1.*/c\CRYST1 261.12 261.12 261.12 90.00 90.00 90.00 P1 1' 6bg.pdb
phenix.fmodel 6bgj.pdb high_resolution=2.04 scattering_table=electron
phenix.mtz2map 6bgj.pdb.mtz include_fmodel=True d_min=2.04 grid_resolution_factor=0.5
1 Like
Ahhh got it - yes that makes sense and agreed
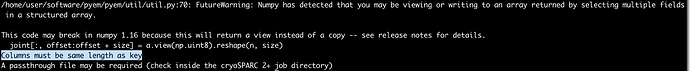
By the way - off topic - do you have any idea why I’m getting this error with csparc2star.py? It was working fine for another job with similar inputs (picked and extracted in cryosparc, J292 is the cryosparc refinement job). Can’t see what is going wrong but I’m sure it is something simple and stupid on my end. The old job still works, so nothing is fundamentally broken in my csparc2star install.
command run: sparc2star.py --passthrough passthrough_particles.cs cryosparc_P36_J292_004_particles.cs cryosparc_P36_J292_004_particles.star
That’s an old version of the program, in the latest one there’s no --passthrough argument (extra .cs files are provided as additional positional arguments) and that error won’t occur. (See the “Final update” thread).
1 Like
aha - got it now - sorry & thanks