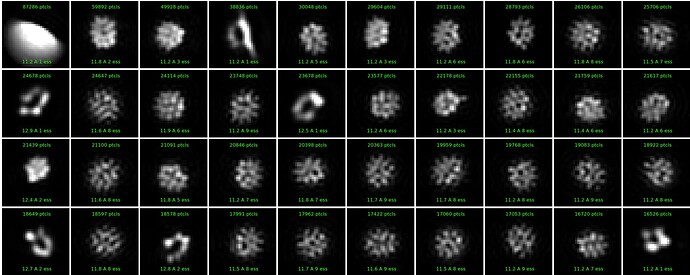
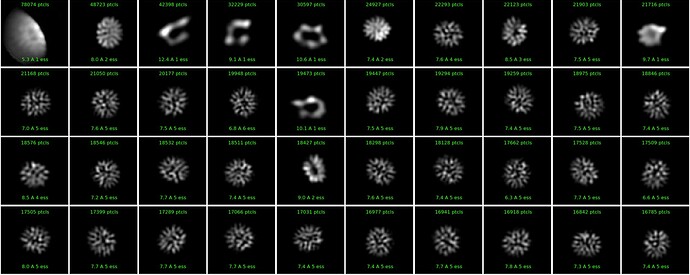
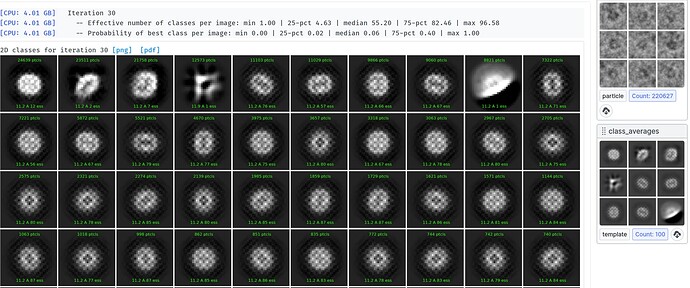
Hi, I see quite strange classes for a small protein (see attached). Tried several options, with- and without forcing poses/shifts, with and without non-negativity forced, with and without clamp-solvent solution, different number of iterations, they all look weird, although particles are well visible in the images. Any idea what could be happening and what else to try?
Thanks,
Michael
Hi Michael,
I had very similar classes with a small transmembrane protein. For me playing around with follwing parameters helped me to get some reasonable classes.
Number of final iterations: 5
online em iterations: 40
Batchsize: 400
Non negativity enforced
Also it helped, in my case, to use a white noise model in the first iterations to get junk and noise classes out. Later I was changing it again to the standard noise model to get higher Resolution 2D averages.
In the end however, I went back to sample/grid preperation as I couldn’t get much out of such datasets.
A few recommended threads in this regard:
Additionally, you could try running your 2D classifcation in Relion - smth I always do with poorly aligning particles. Sometimes I get totally different results there, especially when using the EM algorithm.
Good luck!
Lorenz
Thank you Lorenz, I will try your settings although do not have too much hope that would drastically improve classification. I see classes deteriorate after 4 - 5 iterations. Might try to use just a few iterations if that helps. Thank you for references as well, I do remember seeing those but forgot when.
All the best,
Michael