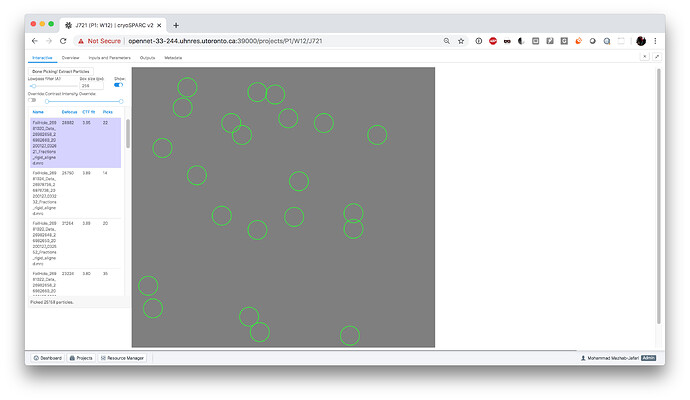
I think I see the issue. Topaz denoise completed successfully and generated outputs, but the outputs are incomplete (only 14 of 29 denoised mics are found in the job dir). Here is the error:
[CPU: 191.4 MB] Using Topaz provided pretrained model.
[CPU: 191.4 MB]
Beginning Topaz denoising command by running command /usr/local/envs/topaz/bin/topaz denoise [MICROGRAPH PATHS EXCLUDED FOR LEGIBILITY] --device 0 --format mrc --normalize --patch-size 1536 --patch-padding 256 --output /home/exx/processing/cryosparc_projects/francesca/P2/J887/denoised_micrographs --lowpass 1 --gaussian 0 --inv-gaussian 0 --deconv-patch 1 --pixel-cutoff 0 --deconvolve --model unet
[CPU: 191.4 MB] Distributing over 4 processes...
[CPU: 186.2 MB] # using device=0 with cuda=True
[CPU: 186.3 MB] # Loading model: unet
[CPU: 186.3 MB] Traceback (most recent call last):
[CPU: 186.3 MB] File "/usr/local/envs/topaz/bin/topaz", line 11, in <module>
[CPU: 186.3 MB] load_entry_point('topaz-em==0.2.4', 'console_scripts', 'topaz')()
[CPU: 186.3 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/main.py", line 148, in main
[CPU: 186.3 MB] args.func(args)
[CPU: 186.3 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/commands/denoise.py", line 547, in main
[CPU: 186.3 MB] , use_cuda=use_cuda
[CPU: 186.3 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/commands/denoise.py", line 292, in denoise_image
[CPU: 186.3 MB] mic += dn.denoise(model, x, patch_size=patch_size, padding=padding)
[CPU: 186.3 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 68, in denoise
[CPU: 186.3 MB] return denoise_patches(model, x, patch_size, padding=padding)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 92, in denoise_patches
[CPU: 186.4 MB] yij = model(xij).squeeze() # denoise the patch
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 476, in forward
[CPU: 186.4 MB] p1 = self.enc1(x)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/container.py", line 119, in forward
[CPU: 186.4 MB] input = module(input)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/conv.py", line 399, in forward
[CPU: 186.4 MB] return self._conv_forward(input, self.weight, self.bias)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/conv.py", line 396, in _conv_forward
[CPU: 186.4 MB] self.padding, self.dilation, self.groups)
[CPU: 186.4 MB] RuntimeError: CUDA out of memory. Tried to allocate 588.00 MiB (GPU 0; 23.69 GiB total capacity; 286.48 MiB already allocated; 27.69 MiB free; 290.00 MiB reserved in total by PyTorch)
[CPU: 186.4 MB] # using device=0 with cuda=True
[CPU: 186.4 MB] # Loading model: unet
[CPU: 186.4 MB] Traceback (most recent call last):
[CPU: 186.4 MB] File "/usr/local/envs/topaz/bin/topaz", line 11, in <module>
[CPU: 186.4 MB] load_entry_point('topaz-em==0.2.4', 'console_scripts', 'topaz')()
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/main.py", line 148, in main
[CPU: 186.4 MB] args.func(args)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/commands/denoise.py", line 547, in main
[CPU: 186.4 MB] , use_cuda=use_cuda
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/commands/denoise.py", line 292, in denoise_image
[CPU: 186.4 MB] mic += dn.denoise(model, x, patch_size=patch_size, padding=padding)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 68, in denoise
[CPU: 186.4 MB] return denoise_patches(model, x, patch_size, padding=padding)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 92, in denoise_patches
[CPU: 186.4 MB] yij = model(xij).squeeze() # denoise the patch
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/topaz/denoise.py", line 510, in forward
[CPU: 186.4 MB] h = self.dec2(h)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/container.py", line 119, in forward
[CPU: 186.4 MB] input = module(input)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/module.py", line 889, in _call_impl
[CPU: 186.4 MB] result = self.forward(*input, **kwargs)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/modules/activation.py", line 714, in forward
[CPU: 186.4 MB] return F.leaky_relu(input, self.negative_slope, self.inplace)
[CPU: 186.4 MB] File "/usr/local/envs/topaz/lib/python3.6/site-packages/torch/nn/functional.py", line 1378, in leaky_relu
[CPU: 186.4 MB] result = torch._C._nn.leaky_relu(input, negative_slope)
[CPU: 186.4 MB] RuntimeError: CUDA out of memory. Tried to allocate 294.00 MiB (GPU 0; 23.69 GiB total capacity; 1.18 GiB already allocated; 29.69 MiB free; 1.43 GiB reserved in total by PyTorch)
[CPU: 186.4 MB] # using device=0 with cuda=True
[CPU: 186.4 MB] # Loading model: unet
[CPU: 186.4 MB] # 1 of 7 completed.
[CPU: 186.4 MB] # 2 of 7 completed.
[CPU: 186.4 MB] # 3 of 7 completed.
[CPU: 186.4 MB] # 4 of 7 completed.
[CPU: 186.4 MB] # 5 of 7 completed.
[CPU: 186.4 MB] # 6 of 7 completed.
[CPU: 186.4 MB] # 7 of 7 completed.
[CPU: 186.4 MB] # using device=0 with cuda=True
[CPU: 186.4 MB] # Loading model: unet
[CPU: 186.4 MB] # 1 of 7 completed.
[CPU: 186.4 MB] # 2 of 7 completed.
[CPU: 186.4 MB] # 3 of 7 completed.
[CPU: 186.4 MB] # 4 of 7 completed.
[CPU: 186.4 MB] # 5 of 7 completed.
[CPU: 186.4 MB] # 6 of 7 completed.
[CPU: 186.4 MB] # 7 of 7 completed.
[CPU: 186.6 MB] Topaz denoising command complete in 57.580s.
[CPU: 186.6 MB] Converting Topaz outputs to cryoSPARC outputs...