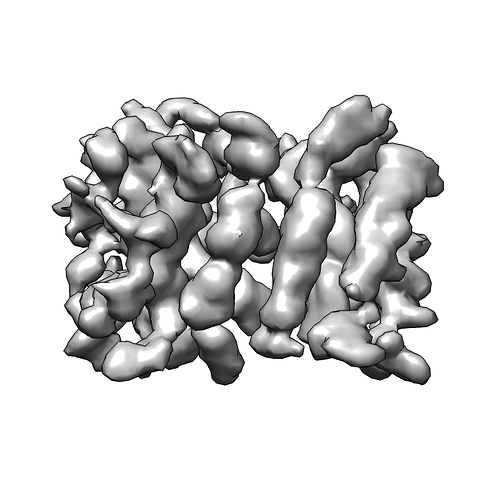
Thanks everyone for all your inputs. I’ve been working on this project for a while and managed to get a structure that is 3.2A after auto-tightening of the mask, but I trust the GSFSC without auto-tightening more (3.4A).
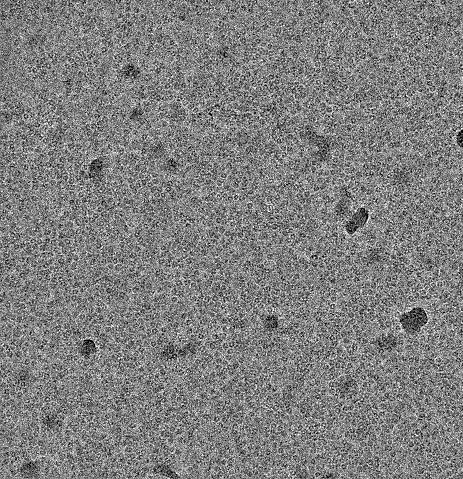
It has been a very tricky project. Not only are particles very small and have low SNR, they are also very densely packed so that signal from adjacent particles is considered during 2D classification. To make things worse, the particle is strongly preferentially oriented. That means that the likelihood of adjacent particles having the same orientation is tremendously increased, leading to 2D classes where the center particle is blurred and adjacent particles resolved.
I took your inputs into consideration. I made sure the particles are perfectly centred and first worked with a box size that is very small (96A for a 60A particle). It was still challenging, but I was able to get good 2D classes. After cleaning the particles, I did an ab initio and heterogeneous refinement of 10 classes. This was important because only one class gave a good reconstruction.
You can see from the graphs how strong the preferential orientation is:
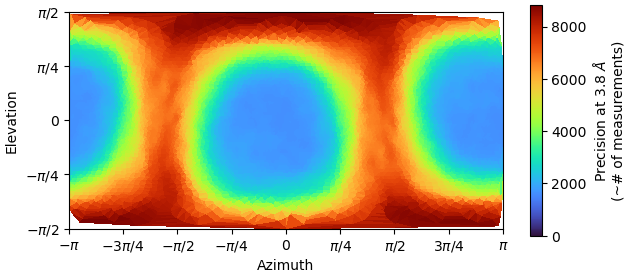
I tried to re-extract at an optimal box size (220A). However, no matter what I did, I could not overcome the problem of adjacent particles’ signal being considered in classifications and reconstructions. I played with myriad of settings and applied very tight masks. But still, the signal from the central particle would be overshadowed by the signal from adjacent particles, so that the central particle lost its features and turned into a blob:
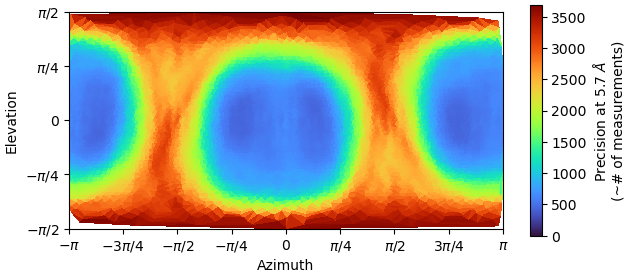
I think the directional distribution precision heatmap is substantiates my point. Working with the same particles, it makes no sense that the directional distribution of particles should change significantly compared to the initial model:
So I went back to the smaller box size (92A) and tried to improve the map via NU-refinement. After analysing the particles of the successful heterogeneous refinement map via 2D classification, I expanded the particle pool. I also generated a mask with relion, which helped a lot in the initial NU-refinement steps. Again, I played with a lot of settings. In the end, what helped was:
- number of extra final passes: 10
- adaptive marginalisation = off
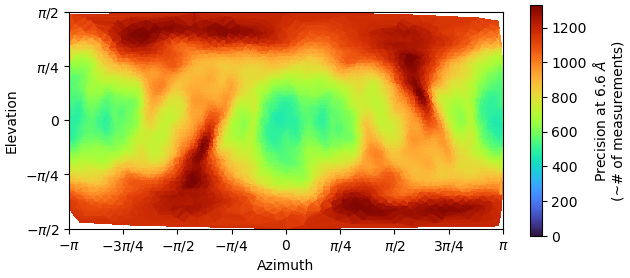
And the directional distribution precision heatmap makes sense:
If you look at the GFFSC after auto-tightening, there is precipitous drop at 4A. Any ideas why this might be?
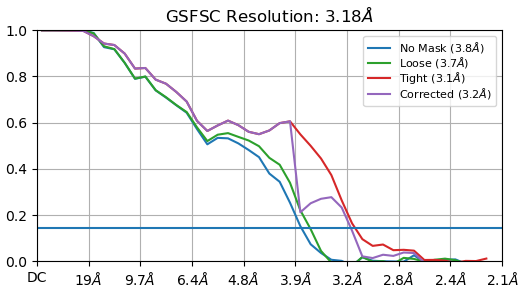
The non-auto-tightened GFFSC looks much better in comparison:
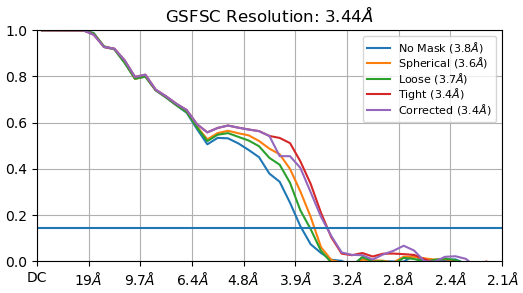
This is the state of things. I can’t use a larger box size. And any optimisation during NU-refinement (defocus, CTF, etc.) actually degrades resolution.
Do you have any inputs or tips (short of acquiring new and better data) how I can make further improvements?
Many thanks for all the help so far and in advance for any future help.