Hi all,
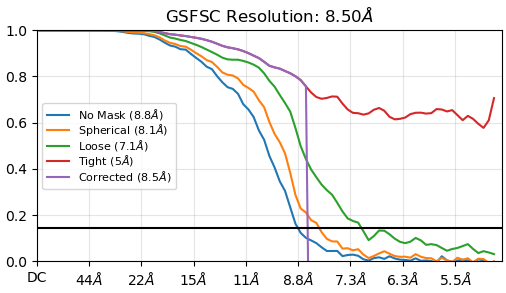
I am assisting a user in processing a medium-resolution data set who sees a very sharp dropoff in the corrected FSC curve around 8.5 A in the non-autotightened curve, although the autotightened curve looks (mostly) fine. It shows up more prominently in non-uniform refinement, but does show up in homogeneous refinement as well. The tight mask appears to be too tight as it does not approach zero.
We’ve tried various things to assess the underlying cause: static masking, no masking (i.e. telling it to start dynamic masking at 1A resolution), very generous dynamic masking parameters, removing duplicate particles, reextracting with a larger box size, doing more final passes, limiting maximum align resolution, CTF refinement, but everything is very consistently showing the drop.
Is this simply where phase randomization occurs? And if so, any ideas why it is so prominent in this data set vs. other data sets? Relevant information may be that this data set is collected at a lower mag / larger pixel size than we use for standard collection (of course I have deleted my pixel size calibration data sets at that pixel size so not sure if it happened there also) and that the dynamic mask when downloaded looks fine but the mask_fsc_auto is indeed quite tight (but unchanged by any parameters we input, I think by design). Also the complex is relatively long and skinny and there are flexible parts, although the reconstruction generally has all the blobs in all the right places.
She’ll be taking a new data set on a higher-voltage scope soon (this was from a Tundra), but since it was so pronounced, just wanted to ask – if the autotightening does indeed seem to almost or fully correct the issue, should we be alarmed by the curve before autotightening?
Thanks as always!
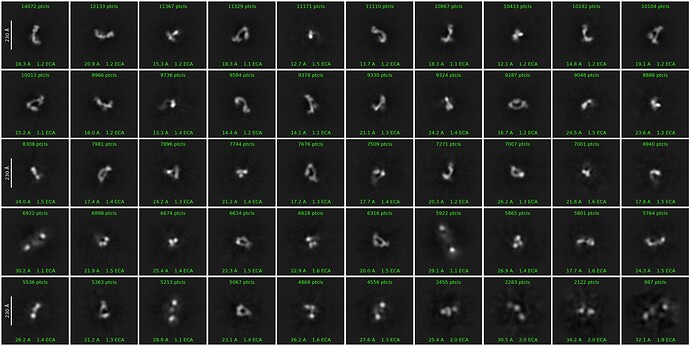
What does 2D look like? Number of particles? As soon as I see an FSC curve behave like that I always think there is a lot of junk hiding… not always, but a fair number of times… particularly with NU refine.
CTF refinement (particularly global) is sketchy at best and dangerous at worst at 5-10 Å.
Magnification should have nothing to do with it, beyond imposing a sampling limit (which can still be broken if you hit Nyquist hard enough…  ) and (at least in my experience) severely miscalibrated pixel calibrations start to cause grief around 4-5 Å…
) and (at least in my experience) severely miscalibrated pixel calibrations start to cause grief around 4-5 Å…
It definitely looks like that is the phase randomisation point, but I don’t think that has anything explicitly to do with the issue you’re experiencing.
Thanks! She’s done a pretty exhaustive classification (multiple rounds of 2Ds, hetero, etc) but the floppiness of the particles means the 2Ds aren’t super clear. 380k particles here:
Do they look like the particles?
Because I’ve seen a couple of datasets over the last year or so where the classes end up looking something like that, but the particles do not look like the classes… and the 3D is absolute garbage.
Yep, they are absolutely the particles, 3Ds look great (although low res but that is expected).
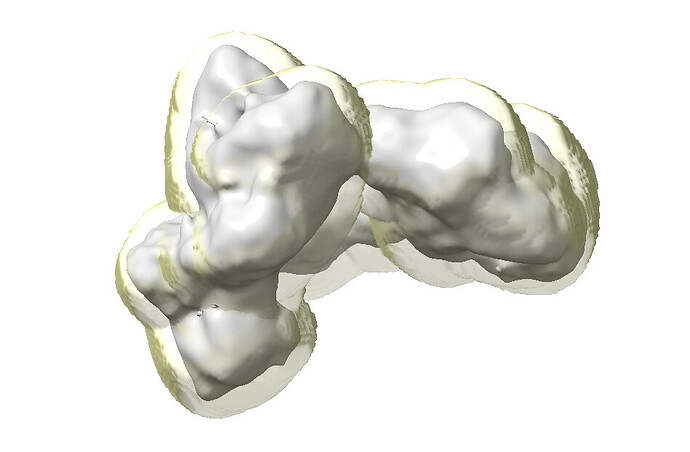
Have you inspected the masks in chimera? Occasionally the automasking routine can generate some odd results
Hi @olibclarke,
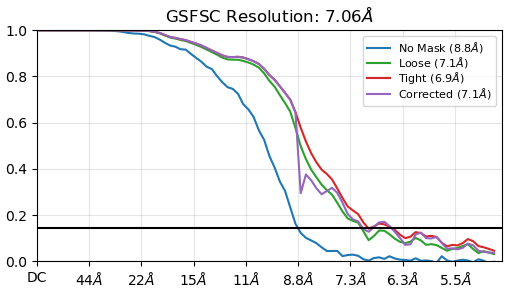
I have looked at the masks, and nothing screamed terrible at me, although I thought the fsc_auto might be tighter than ideal and could be causing the issue (but this does not seem to be addressable by the user so that bias can’t be introduced, was my understanding from whatever documentation I ran across).
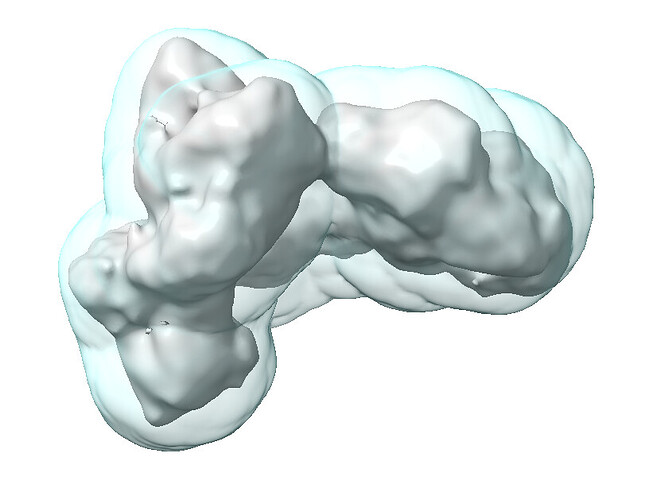
Here’s the CryoSPARC-generated dynamic mask:
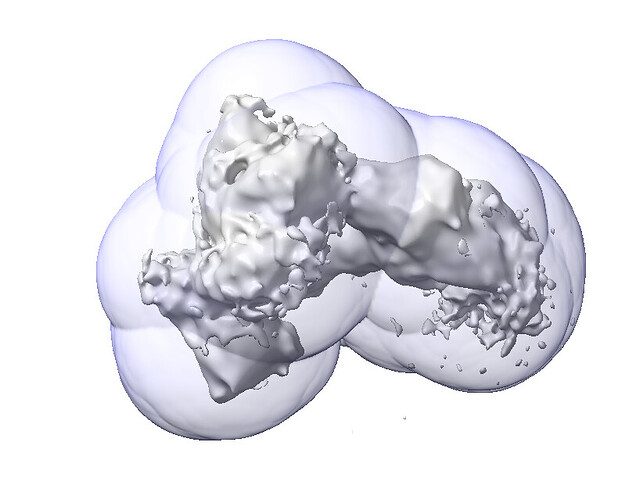
Here’s the fsc_auto for the same refinement:
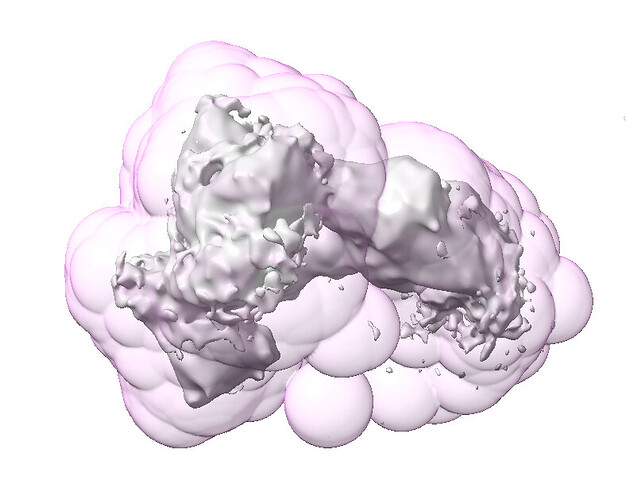
Here’s the same fsc_auto but with the output map at a higher threshold:
I have also tried with much larger user-defined static masks, and no masking at all, but we see the same results (unless I am incorrectly inputting things, but I have tried both giving it a static mask and making “dynamic mask start resolution” 1) and we see the same behavior. Here’s two examples of static masks we tried supplying:
I even got paranoid that for some reason it wasn’t applying the static mask (or no mask) that we were supplying it, but it does seem to be doing that correctly: