Hi, cryoSPARC community,
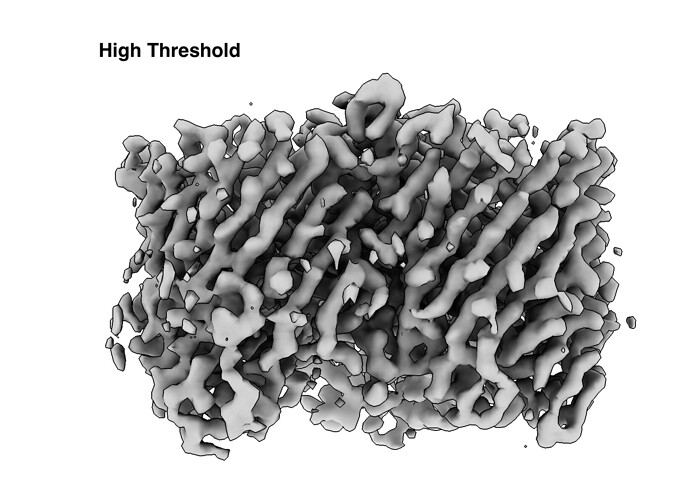
I’m working on resolving a protein complex structure of two proteins, one an outer membrane protein associated with DDM micelle and its binding partner. Since I’m working with membrane proteins, I used the following scheme: ab initio reconstruction (two different jobs: 1 class and 3 classes) and non-uniform refinement. Using non-uniform refinement I have resolved a nice ~3ish Å resolution map of one of the proteins, but its binding partner appears to be more flexible and at a low threshold appears to have a density at the same level of noise. When the threshold is increased to remove the noise density from the binding partner also disappears (example shown below). From what I have read non-uniform refinement is best at resolving regions of flexibility, but this doesn’t seem to be the case in my situation. I’m hoping you all may have suggestions I have not tried yet. So far, I have tried particle subtraction, subtracting the outer membrane protein’s density, local refinement, and different masking strategies, but have had no luck.
Any suggestions or insight is much appreciated. Thank you, Leti