Hi!
I saw the post 3D reconstruction of simulated particles with high SNR - 3D Reconstruction - CryoSPARC Discuss, and tried to do reconstruction from clean, no-CTF, simulated projections, but got wrong 3D map result. I’m working with CryoSPARC v4.0.2, and my workflow and 3D map result are shown in the following figure.
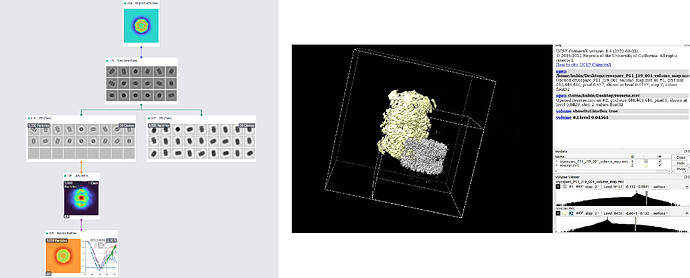
The left figure shows the workflow.
-
Import Volume: The imported volume is the result of Homo Refine job in the cryosparc T20S tutorial. (Done in advance, and the 3D map result is good, and 9378 particles are contributed to the final map reconstruction.)
-
Simulate Data: I set SNR=99999999 and turn off Apply CTF to images to simulate 9000 clean projections. Other parameters are as default.
-
2D Class: As for simulate data, I think 2D class is not necessary, but according to the post link above, I do the 2D Class to make cryosparc ignore CTF. In the right one (J17), all parameters are as default; while in the left one (J16), I turn off Do CTF correction to make cryosparc ignore CTF in following processing.
-
Ab initio: All parameters are as default.
-
Homo Refine: I give D7 symmtry and other parameters are as default.
I open the result 3D map in ChimeraX, as shown in the right figure. The gray one is the 3D map result of Homo Refine job. It’s absolutely wrong, not like T20S structure at all. I suspect it’s because something wrong with blob/sign, as 2D Class result is not so right. So, I reverse the 3D map, that is, read the .mrc map file with mrcfile python package, and multiply all elements with -1, and save the map. The reversed map is the yellow one in ChimeraX. It looks more like T20S than the gray one, but is also absolutely wrong compared to the real T20S structure. (Besides, after open two .mrc map file in ChimeraX, the yellow one looks bigger than the gray one, and I don’t know why.)
As I use clean projections to reconstruct, I expect to get a good 3D map result, but it turns out not the case. I’m wondering what’s wrong or why is this. Anyone has any advice?
Best,
Ciren
Hi @CirenSangzhu,
In CryoSPARC v4.1 we fixed a bug that caused the simulator to not write the correct CTF metadata when “Apply CTF to images” was disabled (see our changelog here). This explains why 2D classification only produced expected results when the “Do CTF Correction” parameter was disabled. If you could update to any version of CryoSPARC later than v4.1, you should see that this workflow works well — please let us know if that resolves the issue!
Best,
Michael
2 Likes
Thanks for your response!
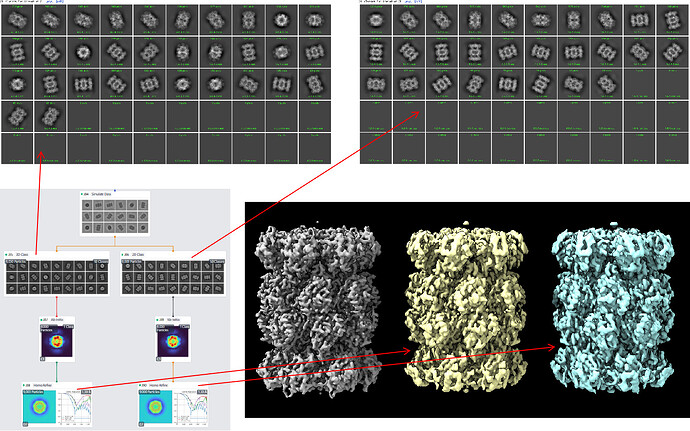
I take your advice and update my CryoSPARC. Now I’m working with CryoSPARC v4.1.0. I repeat previous operations in the new version, as shown in the following figure.
Parameters are the same as previous. I generate 9000 simulated clean projections (SNR set to 99999999 and Apply CTF to images turned off). After updating, 2D classification job seems doing well either with Do CTF correction turned off (J85) or turned on (J86).
However, the final map after Homo Refine job is still not satisfying. As shown in the figure, there are three maps opened in ChimeraX. The gray one is the original one, which is imported to be the input of Simulate Data job. The yellow one and blue one are reconstructed using simulated clean projections. As you can see, both the yellow map and blue map are worse than the original gray map.
In my understanding, simulated clean projections have better quality compared to original particles. They do not have noise in images, and are not corrupted by CTF, so the reconstruction should result in better map, or at least be as good as the original gray map. But it turns out not the case. I don’t know why. Can you help me?
What’s more, as you can see, the FSC curve is unusual. The reported resolution is 1.33 angstrom, but I think the quality of reconstruction maps are not such good.
I also check the .cs file of simulated clean particles, as @mmclean suggest there is a bug writing wrong CTF metadata to .cs file in older version. I check the CTF parameters in .cs file now, and I think these parameters lead to constant CTF at -1, not 1. Is this a bug, or intentionally designed? Because I think it clean projections should have CTF equals to constant 1.
Thanks,
Ciren
Hi @CirenSangzhu ,
The CTF being constant at -1 is intentional, and is related to the sign convention of the particle images relative to the 2D references.
Could you post screenshots of the volume used in the simulate data job (just the Fourier space slice plots, from the upstream job from which it was connected to the simulator job), as well as screenshots of the Fourier slice plots from the final homogeneous refinement? Also, could you indicate what the GSFSC resolution of the input volume is? The Homogeneous Refinement maps’ FSCs from synthetic data may be sensible depending on the actual resolution of the input reference. The input reference is low-pass filtered to the GSFSC resolution and this means fourier coefficients beyond this resolution have near 0 amplitude, which means the synthetic data has near 0 amplitude fourier coefficients at high-res, meaning the FSC will not be meaningful beyond the GSFSC.
Regarding the map quality, what you’re observing may also be expected: you shouldn’t ever observe the reconstructed map from clean simulations looking better than the initial map. The amount of signal in the reference is fixed and synthetic slices can’t create more signal than is other present. In general, maps from synthetic data will look lower resolution than the initial reference because there are other factors such as orientation distribution of the synthetic slices that might not be optimal. If you want to make the synthetic data as high quality as possible, you can repeat simulation while:
- make the orientation sampling more thorough by increasing the “Number of gridpoints for rotations” to at least 40,
- creating more slices (>20k)
Then, during refinement you could enforce the correct D7 symmetry during homogeneous refinement. If you then take the output map from refinement and lowpass filter it via Volume Tools to the GSFSC resolution of the original experimental reference that was input to the simulator, you may observe maps with similar features to the original map.
Best,
Michael