Thank you both for all of your insightful comments and the time spent on them!
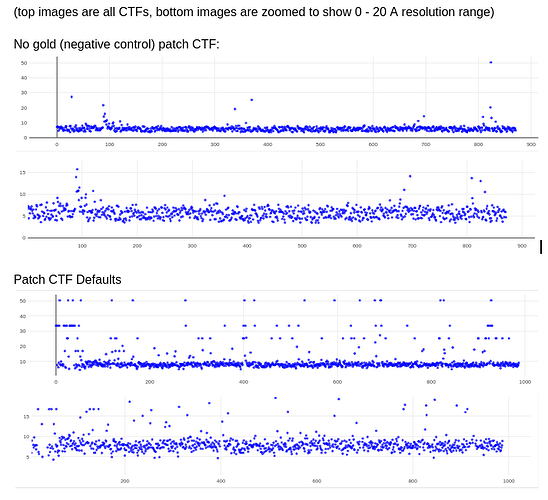
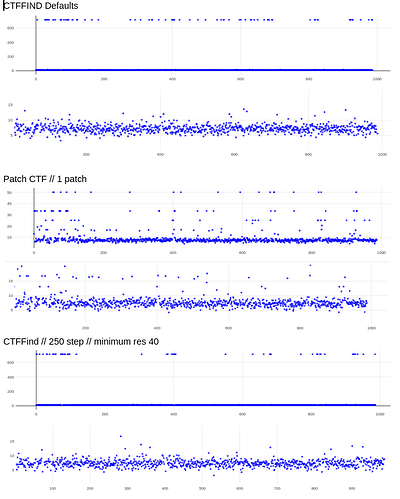
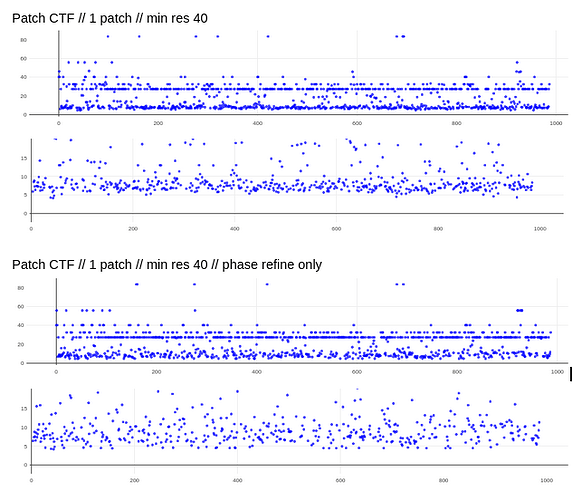
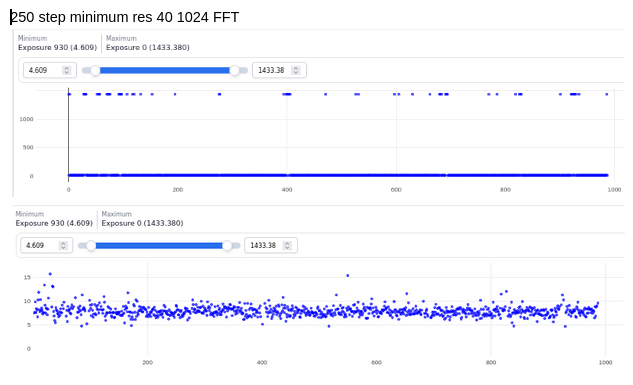
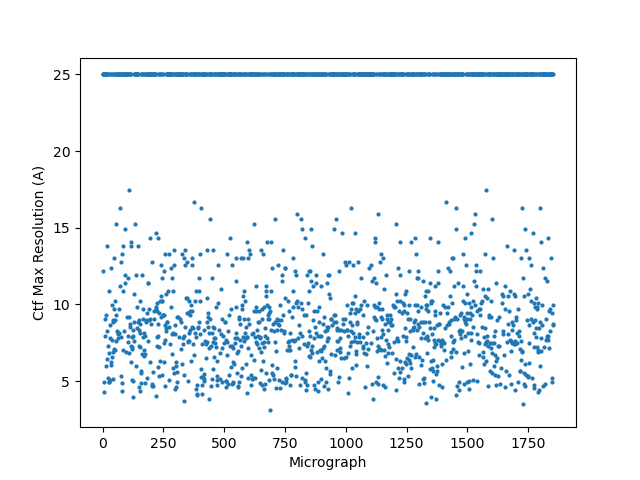
@DanielAsarnow I see what you are saying re: 20A still being too high in those cases, thanks for making that clearer for me. I tried running Patch CTF, rejecting everything that fails badly (honestly we often have our limit around 10-12A anyways), and then running CTFFIND4 on the rest – it saved about half of my micrographs that were rejected. These results are already running in exhaustive mode, except for the defaults run.
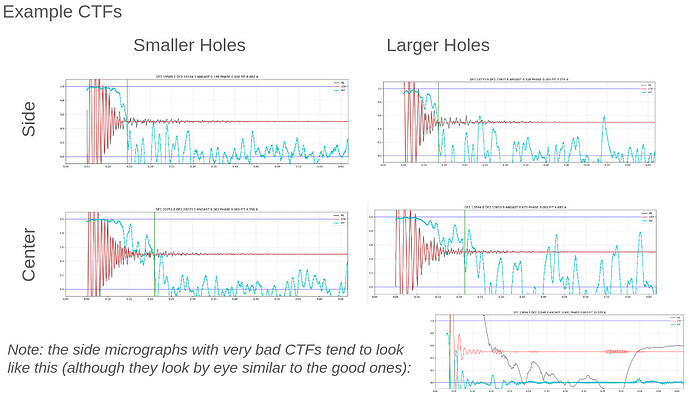
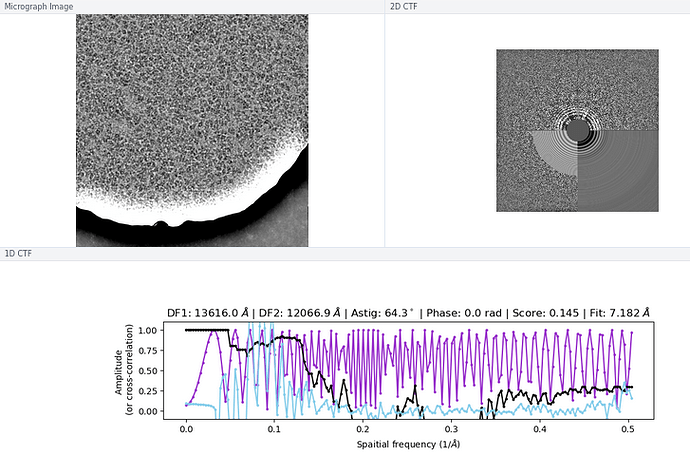
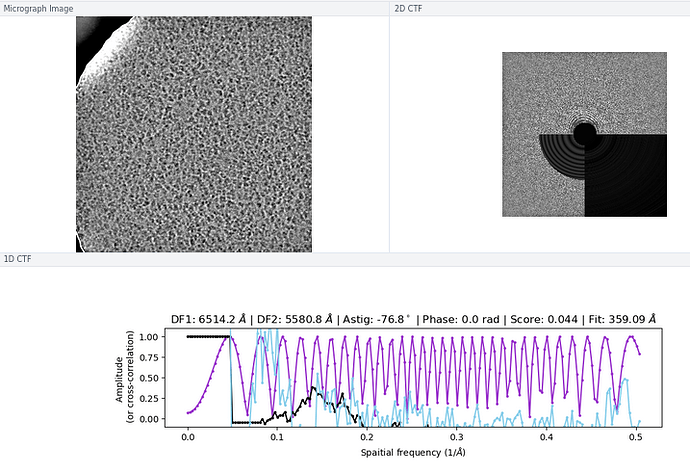
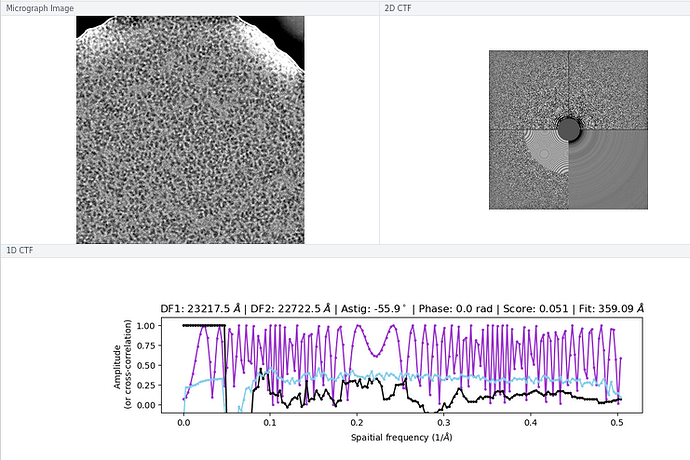
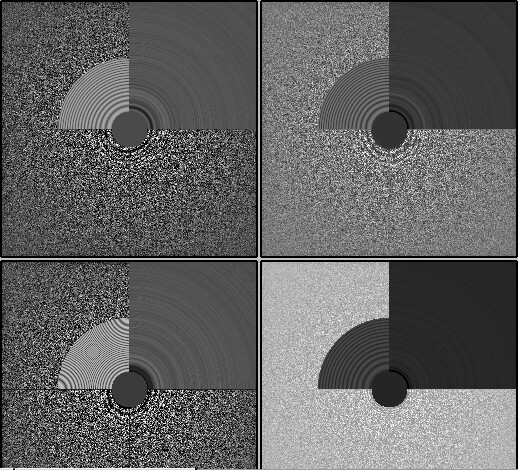
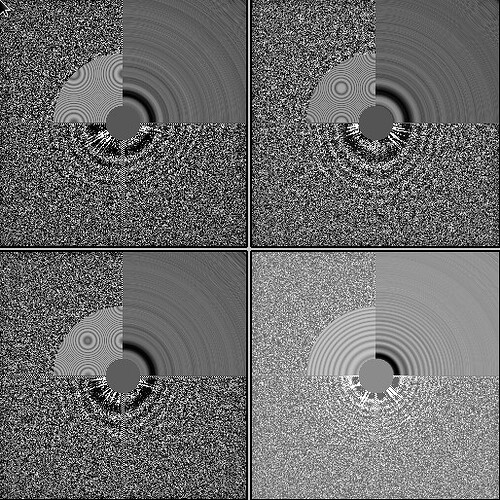
My comment around the empirical FFT was that the Thon rings appear much clearer in the top example than in the other two to me by eye (and in the radial average). Makes sense that it could be a function of the background and/or where the cutoff is. I suppose the amount of work that should go in to saving some of these micrographs is dependent on how painful it is to generate more images and what fraction can be discarded, but it seems at least that windowing / masking out gold definitely turns some of these data sets from unuseable to mostly useable.
@rbs_sci Thank you for the input on loosening astigmatism parameters if needed – I do think any astigmatism problems we are experiencing are pretty secondary to the CTF issues. We’ll keep it in mind should more primary astigmatism issues arise.
I have tried doing CTF refinement before on some of the particles from “bad” data sets, but I think the original fit was poor enough that it wasn’t saveable, so the lowest resolution few peaks fitting is good to keep in mind. Also, I’m a huge fan of the CTF Simulator, so thanks for posting and I will take a look in this particular case re: FFT size.
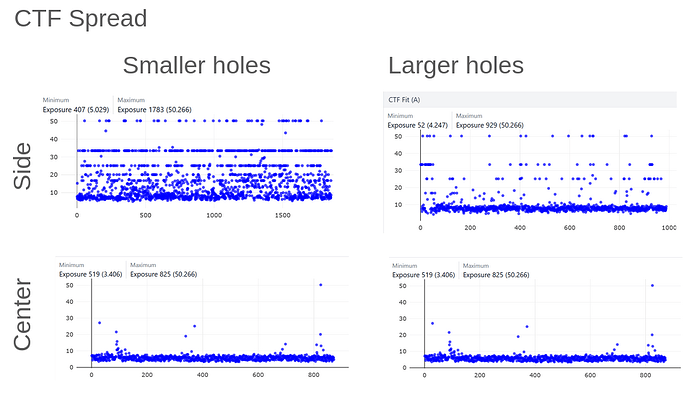
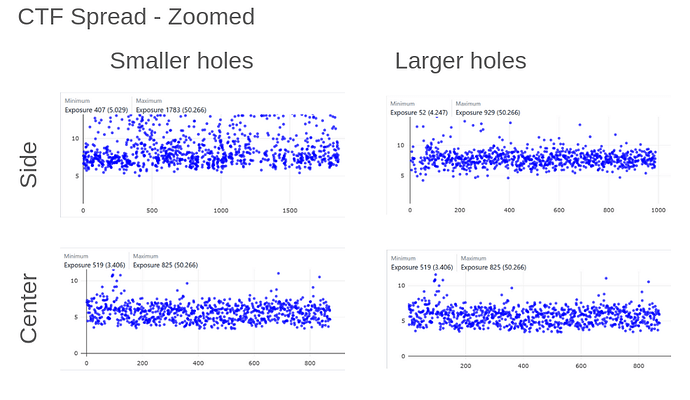
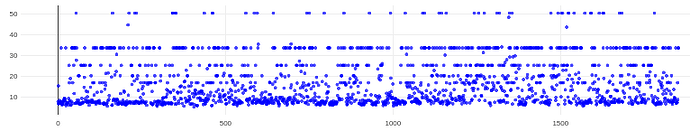
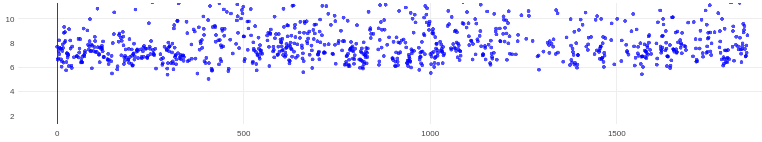
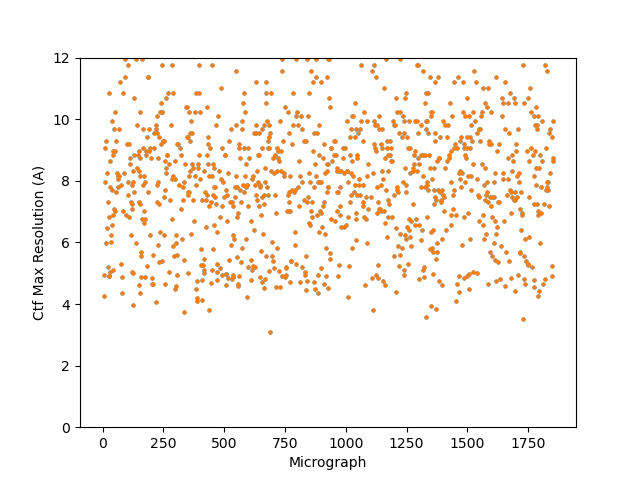
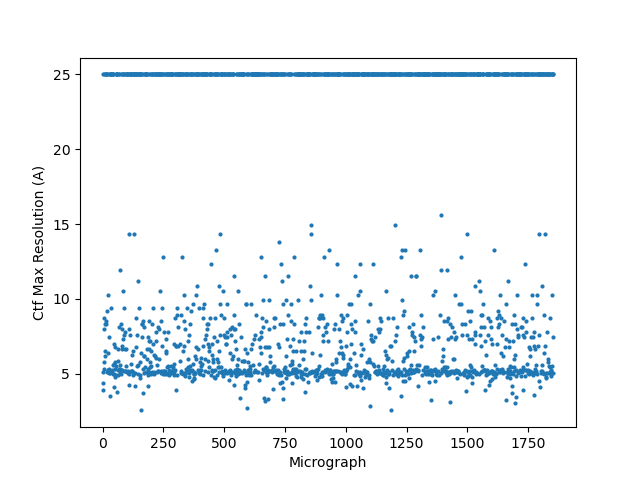
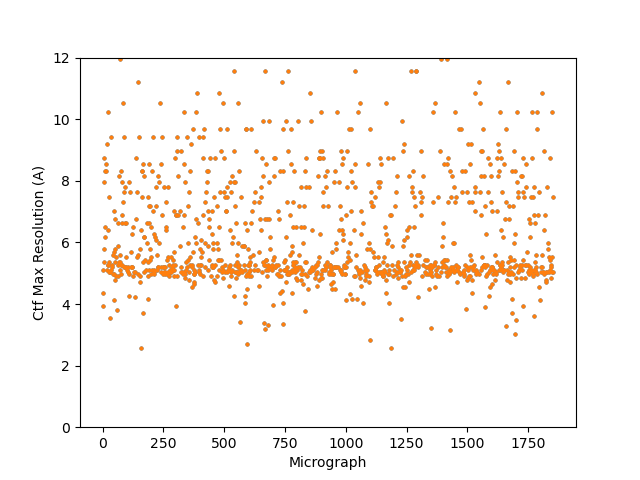
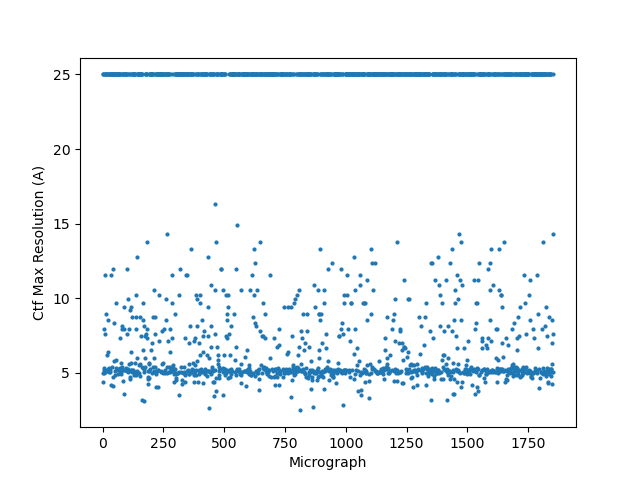
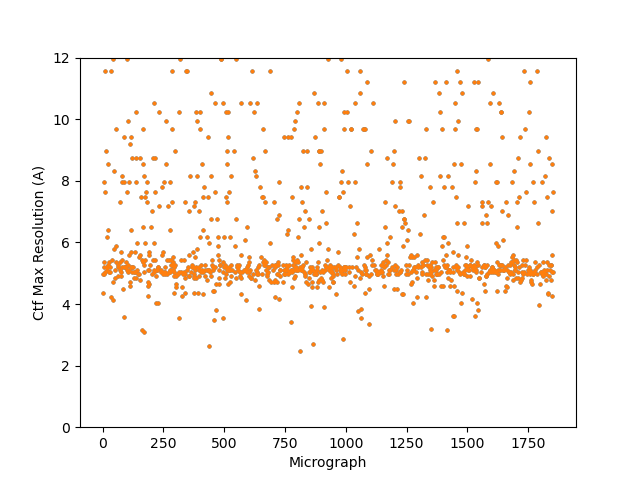
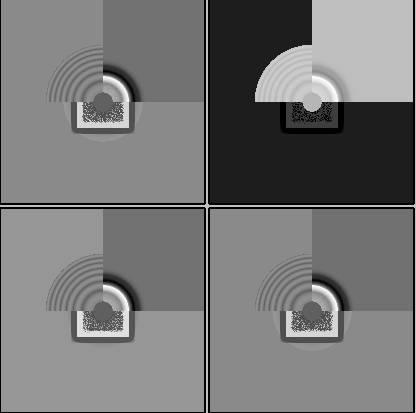
On the Relion windowing size issue: Good catch @DanielAsarnow, I did mean average frame PS. Dumping in the micrographs from a default MotionCorr job (which are still 4096x4096) and then specifying “yes” to “use power spectra from motioncorr job” requires that you specify the window on the scale of 512x512. Otherwise you will get empirical data that look like QR codes  (the scale also becomes very off; below the “yes” images are shown at a scale of 0.1 and the “no” images are shown at a scale of 1). If you say “no” then you can use the full size, i.e. 2048, and the outputs look reasonable. I’m a little hazy on where windowing comes in post-PS calculation, but using the motioncorr PS vs not seemed to “save” slightly fewer micrographs but overall give lower average estimated CTFs (see below; I guess it is a trade off between more data and the gold reflections but still not sure where windowing comes in if the PS are already calculated?).
(the scale also becomes very off; below the “yes” images are shown at a scale of 0.1 and the “no” images are shown at a scale of 1). If you say “no” then you can use the full size, i.e. 2048, and the outputs look reasonable. I’m a little hazy on where windowing comes in post-PS calculation, but using the motioncorr PS vs not seemed to “save” slightly fewer micrographs but overall give lower average estimated CTFs (see below; I guess it is a trade off between more data and the gold reflections but still not sure where windowing comes in if the PS are already calculated?).
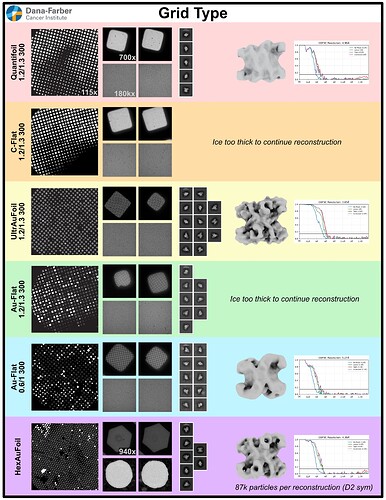
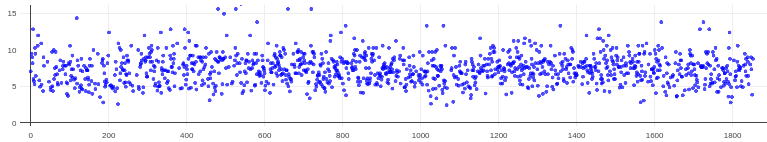
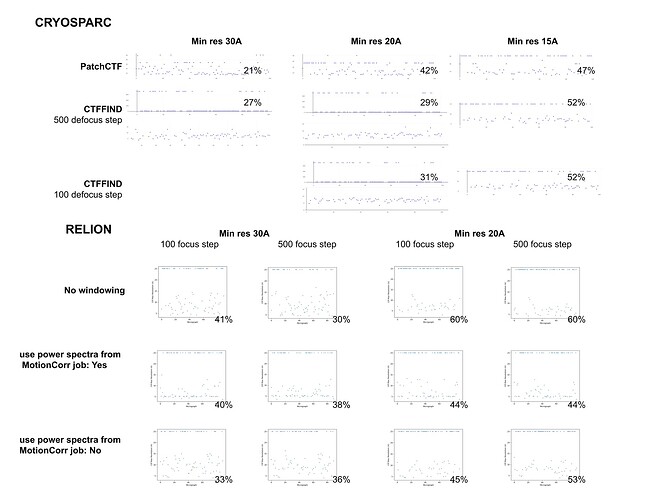
Hopefully anyone interested (I can’t resist an opportunity for a good side-by-side comparison) can zoom in on this image – the percentages are the percent of micrographs that were assigned an extremely high CTF max res (generally >30A but otherwise the upper ceiling).
Probably the take away here is there isn’t one “fix” (except maybe gold masking??) and the best thing is, as @DanielAsarnow suggested, to process different micrographs in different ways depending on how the results of the initial rounds of patchctf and CTFFIND look. For us the windowing does seem to mostly rescue much of the data set to where it would be expected if there were no gold in the image, so if anyone gets stuck here in the future, that’s where I would start, but we’ll be trying to avoid much gold in the image if at all possible.
All good suggestions and I appreciate the discourse – hoping that as CryoSPARC continues to advance there will be the option to process these sorts of difficult micrographs using windowing/masking within the interface!
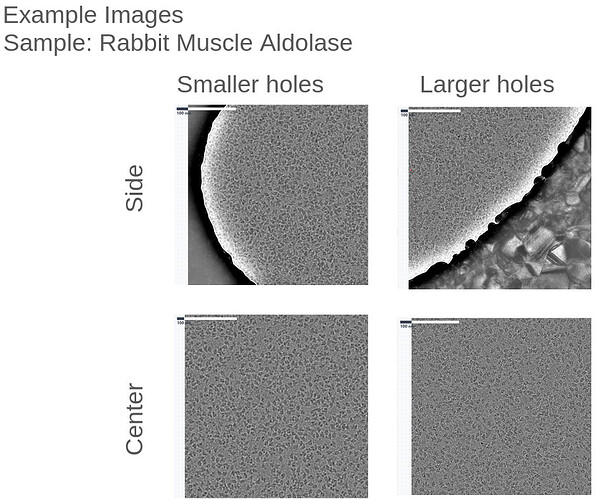
![]() ). Everything was collected on the Tundra with a Ceta-F at 0.748 A/pix (306x306 nm images).
). Everything was collected on the Tundra with a Ceta-F at 0.748 A/pix (306x306 nm images).