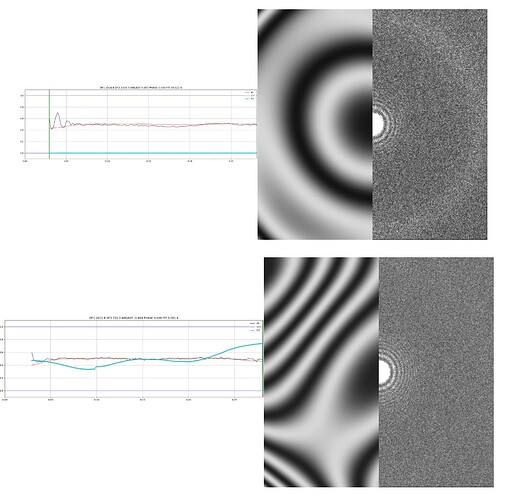
We’ve collected a small dataset, and the micrographs (gold grid) look promising; we can even see the shape of the particles. However, after processing them with CryoSPARC, the CTF results are poor (see attachment). I mostly used the default parameters and didn’t make many adjustments. Does anyone have any suggestions for improving the results? Thanks!
Use a single patch (Override x/y knots to 1), minimum defocus to 30 or 40… or if you have 32-bit MRC micrograph output (not too demanding to use if dataset small and/or need to redo) use CTFFIND with exhaustive search and defocus step 250 Å - it’s slow but in the past has given me good CTF fits from micrographs which everything else fails with.
edit: If aliasing visible, might need 1024 FFT in CTFFIND as well.
could this be pixel input error? something to double check.