Hi, I have been using “Minimize over per-particle scale” in Local Refine New, which seems to work well for my merged datasets.
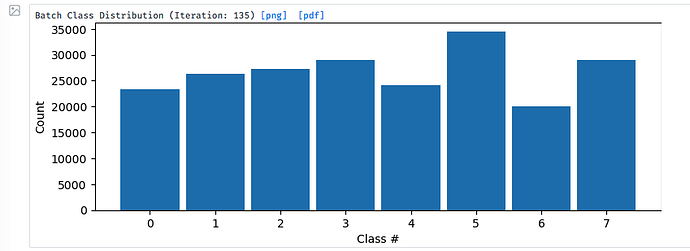
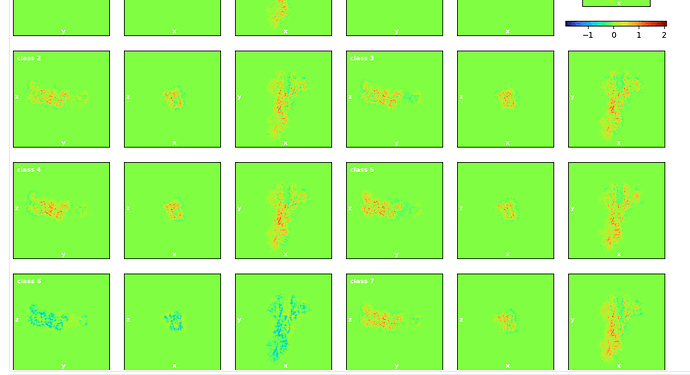
However, the following 3D classification beta revealed something new: there is always one class (class 6) with negative density values (see the attached picture), whose distribution seems to match the real density distribution as reflected by other classes. A follow-up Local Refine of this peculiar class 6 produced a very noisy reconstruction.
I wonder what process could produce such negative densities. Thanks!
Probably junk particles. Have you tried 2D classification of this subset of particles?
Great suggestion! Let me try it.
If they are junk particles, how come their negative densities perfectly match the real densities?
If they are junk particles, how come their negative densities perfectly match the real densities?
Because you are classifying without alignment - the orientations of the particles are unchanged and match the consensus reconstruction. I suspect if you run ab initio or 2D on these particles you will get garbage
Yes 2D classification of particles in class 6 did produce garbage 
Thanks for the explanation!
@zhangrui_wustl may I ask you if you used the default parameters in your 3D classification, and if you supplied a mask too ? Thanks
Yes, mostly default parameters in 3D classification with PCA option.
I supplied a mask that covers the whole complex.
Hey! Is this a new way to get rid of junk particles? Sounds interesting…
Hey @jenchem, yes definitely – junk removal was one of our imagined use cases for 3D classification.
@zhangrui_wustl thanks for your answer. So you do not supply any volume, correct ?
Correct. You only provide volumes for supervised classification.
Hi @vperetroukhin I have been playing with 3D classification beta for a while, so far the results in my hand are not as good (e.g. clean separation) as Class3D in Relion3 with alignment off (sorry for the subjective statement  , just want to give you some feedback as one data point).
, just want to give you some feedback as one data point).
In Relion3 Class3D one can tune the T number between 4 and 100. Is there similar parameters in cryosparc 3D classification?
@zhangrui_wustl no worries, thanks for the honest feedback! We’re actively working on improving the job. Two thoughts:
-
As discussed in this thread, please keep in mind that the 3D Classification job uses weighted back-projection for the 3D density outputs. You may see ‘cleaner’ separation if you do a homogeneous reconstruction or refinement on the individual particle sets.
-
There is nothing directly analogous to the RELION T parameter, though you can adjust both the
Target resolution(which will filter out higher frequency components), theInit structure lowpass resolutionandTarget class ESS fraction(which can make the initial class distributions more uniform).
Valentin
@vperetroukhin Thanks for the explanation!
Just to clarify, by “not as good” I mean the 3D classification results by CryoSPARC (using PCA option) tend to look more ‘similar’ to each other even after examining subsequent local refinement of each class; while in Relion3 I could get more distinct good-looking and bad-looking classes.
I have been setting Target resolution to 6 Å or 8 Å, do you recommend 15 Å or 20 Å if I am looking for more global variations?
I am running 3D classification beta with nearly 500K particles and NU refinement as input, and particles were extensively cleaned up with 2D classification. I have indicated 6 classes. It takes 3.5 days to finish in simple mode. With PCA option one iteration takes 1 hour (there are more than 90 iterations in total). Is there any parameter one could modify to have 3D classification running faster ? It’s running on an 11 GB card.
Many thanks
I have been setting Target resolution to 6 Å or 8 Å, do you recommend 15 Å or 20 Å if I am looking for more global variations?
@zhangrui_wustl yep this is possible, but global variations like that will probably be better analyzed using the heterogeneous refinement job (where alignments can also be refined).
I have indicated 6 classes. It takes 3.5 days to finish in simple mode. With PCA option one iteration takes 1 hour (there are more than 90 iterations in total). Is there any parameter one could modify to have 3D classification running faster ? It’s running on an 11 GB card.
@marino-j The PCA/simple options refer to the initialization of densities, so this shouldn’t affect the per-iteration time. You can take a look at increasing the Batch size per class which will reduce the amount of online-EM iterations required for one epoch.