To follow up on this, here is an example (using the ryanodine receptor as a test case).
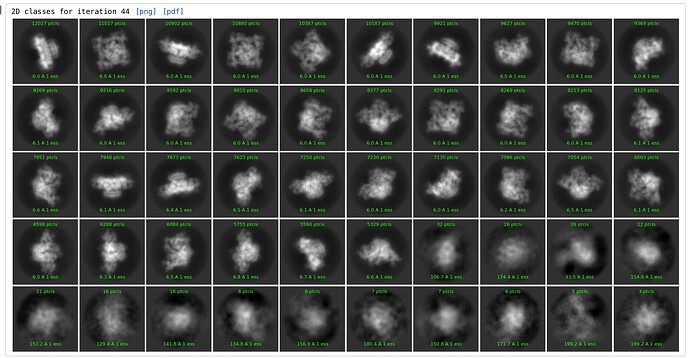
Defaults: Start sigma annealing at it=2, for 15 iterations, 40 O-EM iterations, 5 full iterations:
All “bad” classes are essntially empty.
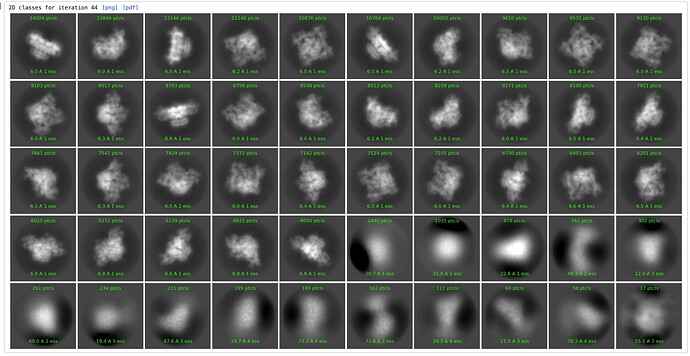
No sigma annealing - Set sigma annealing start iteration to 200, effectively switching it off; other parameters unchanged:
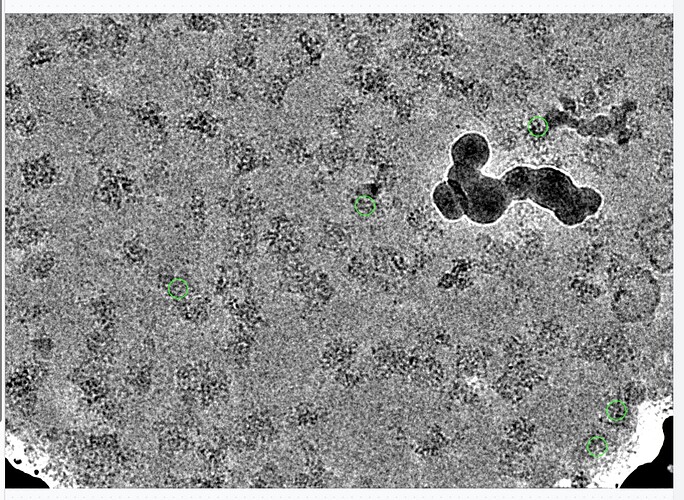
There are now several thousand particles in bad classes. These particles are in fact bad, as we can see looking at particles or micrographs, they are all next to gold or junk:
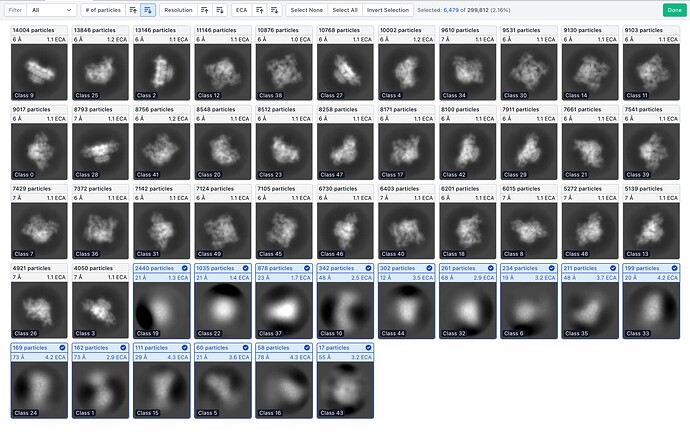
In this case, another round of 2D with sigma annealing switched off, and recentering switched off, allowed identification of an additional set of bad particles, which were otherwise subsumed in the good classes.