So I’ve been getting this extra bump (around 6.5 Å) in my FSCs from a variety of programs for my dataset.
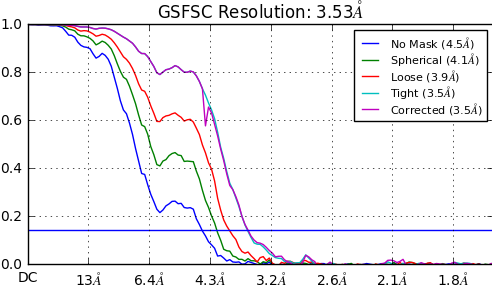
I’ve seen this (GSFSC curve bilobed) and this (Tight, corrected and loose GSFSC curves) and Penzcek’s chapter about common FSC issues (https://books.google.com/books?id=D1PHZBdDkjEC&pg=PA98&lpg=PA98&dq=fsc+artifact+cryoem&source=bl&ots=CImN2pr1Wt&sig=d2SD2nrPH36s1Gc5UQ9JMjiMc6Q&hl=en&sa=X&ved=0ahUKEwi38av0gNzSAhVG9mMKHam4BhgQ6AEISjAG#v=onepage&q=fsc%20artifact%20cryoem&f=false)
Any ideas on why I see this bump? One of the links suggests crowded micrographs with overlapping boxes could cause this. I had a look at the FSC of the super crowded aldolase paper from Lander lab (https://www.biorxiv.org/content/biorxiv/early/2017/05/25/141994.full.pdf) and it doesn’t have a bump like this. I’ve checked to ensure I don’t have overlapping particles, but my micrographs are crowded. I suppose the issue could be related to estimating the noise, since some of the background in my boxes is other particles?
I can confidently assign alpha helices, in my density, so is this type of FSC still acceptable? I’ve use ab initio models to refine the dataset.