Dear all,
I was able to import the beam shift/image shift values for a dataset collected via Leginon/Appion and wanted to share here.
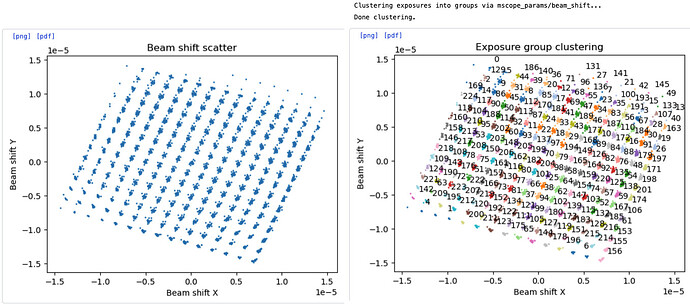
In line with what @mmclean explained in this topic and in another post, I generated XML for each micrograph in the dataset, imported the beam shift values in the XML files via Import Beam Shift, and ran the clustering & exp_group_id mapping procedures via Exposure Group Utilities (gave both the exposures and particles as input).
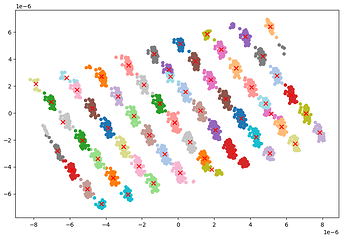
The image above shows that agglomerative clustering method can cluster 225 exposure groups for this particular dataset.
To generate XML files, I first prepared the data (micrograph name, image shift X & Y) obtained from Appion as a CSV file called mics_imageshift.csv. The CSV format follows:
micrograph1.mrc,-0.00000022151753017022197,-0.0000003004722777211951
micrograph2.mrc,-0.000006418716276264378,-0.0000032146108280230503
...
Next, we just need to write the XML files for each line in the CSV list.
Here’s a python script to generate XML reading data from CSV. lxml is required to run the script (pip install lxml):
#!/usr/bin/env python3
import csv
from lxml import etree
import sys
import argparse
def update_xml(xml_content, x, y):
try:
root = etree.fromstring(xml_content)
for element in root.xpath('.//a:_x | .//a:_y', namespaces={'a': 'http://schemas.datacontract.org/2004/07/Fei.Types'}):
element.text = str(x) if element.tag.endswith('_x') else str(y)
updated_xml = etree.tostring(root, encoding='utf-8', xml_declaration=False).decode('utf-8')
return updated_xml
except Exception as e:
print(f"Error updating XML: {e}")
return None
def generate_xml_from_csv(input_csv):
try:
with open(input_csv, newline='') as csvfile:
csv_reader = csv.reader(csvfile)
for idx, row in enumerate(csv_reader, 1):
filename, x, y = row
xml_content = """
<MicroscopeImage xmlns="http://schemas.datacontract.org/2004/07/Fei.SharedObjects">
<microscopeData>
<optics>
<BeamShift xmlns:a="http://schemas.datacontract.org/2004/07/Fei.Types">
<a:_x></a:_x>
<a:_y></a:_y>
</BeamShift>
</optics>
</microscopeData>
</MicroscopeImage>
"""
updated_xml = update_xml(xml_content, float(x), float(y))
if updated_xml is not None:
with open(f'{filename[:-4]}.xml', 'w') as xmlfile:
xmlfile.write(updated_xml)
else:
print(f"Skipped XML {idx}: {filename[:-4]}.xml")
print("Done generating XML files!")
except FileNotFoundError:
print(f"Error: File '{input_csv}' not found.")
sys.exit(1)
except csv.Error as e:
print(f"Error: reading CSV file '{input_csv}': {e}")
sys.exit(1)
except Exception as e:
print(f"Unexpected error: {e}")
sys.exit(1)
def main():
parser = argparse.ArgumentParser(description="Generate XML files from a CSV file.")
parser.add_argument("input_file", help="Path to the input CSV file")
args = parser.parse_args()
if args.input_file.lower().endswith('.csv'):
generate_xml_from_csv(args.input_file)
else:
print("Error: Unsupported file format. Please provide a CSV file.")
sys.exit(1)
if __name__ == "__main__":
main()
I saved the script above as csv2xml.py and ran the script as:
./csv2xml.py mics_imageshift.csv
The script will write micrograph1.xml, micrograph2.xml, … in the same directory.
With the generated XML files, we can now import beam shift with movies/micrographs from the start of processing pipeline or import beam tilt for micrographs and particles that have been processed before v4.4.
Kookjoo