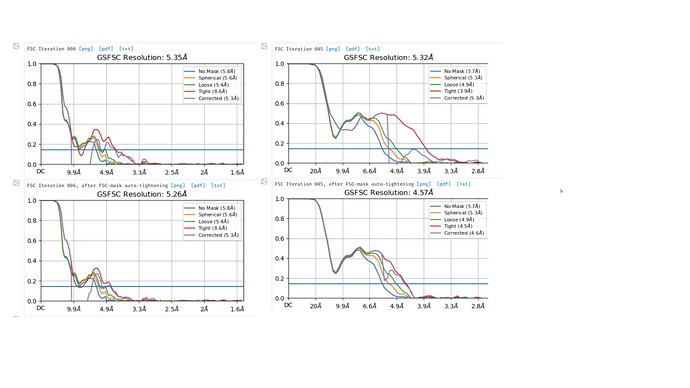
Hello:
I am currently working on a small ectodomain of a protein. And started to do homogeneous refinement after multiple rounds of 2D classification. However, my GSFSC graph always have the a dip around 10~5 angstrom. I have looked into the forum to see how people with similiar situation addresses the problem. I have tried scaling up and down my box size in extract from micrograph, importing a static mask and altering my dynamic mask threshold but neither of them really helped and the GSFSC graph still have a large dip.
Thanks