Hi @yurotakagi, no worries. Let’s try to get it working on your macbook, since that way we don’t have to worry about permissions. Here is how I would install cryosparc-tools on my mac:
1. Install miniconda
If you already have conda installed, skip this step
Following the instructions here (Installing on macOS — conda 24.1.3.dev60 documentation), install miniconda.
2. Create a conda environment
Generally, it is best practice to use virtual environments with python. This helps avoid conflicts if certain packages need specific versions of other packages.
I will use this command to create a python environment called cs-tools:
conda create --name cs-tools python=3
3. Install cryosparc tools and other libraries
We must activate the virtual environment before installing packages or using cryosparc tools. One way to check which environment you’re in is conda info --envs:
$ conda info --envs
# conda environments:
#
base * /Users/rposert/miniconda3
cs-tools /Users/rposert/miniconda3/envs/cs-tools
Right now, I’m in my base environment. I do not want to install packages there! If I first activate my cs-tools environment:
$ conda activate cs-tools
we can see that I am now in the correct environment:
$ conda info --envs
# conda environments:
#
base /Users/rposert/miniconda3
cs-tools * /Users/rposert/miniconda3/envs/cs-tools
Note that the * is now next to the cs-tools path instead of the base path.
We can now install the packages we need! In addition to the packages that cryosparc-tools requires, I find these packages useful when writing scripts and inspecting data:
pandas, for inspecting data as a data framematplotlib, for simple plots
so we can install the packages to the virtual environment like so:
$ python -m pip install cryosparc-tools pandas matplotlib
Now the environment is ready to go!
4. Set up SSH tunnels
We now need to set up some SSH tunnels so that our computer can connect to the cluster which hosts CryoSPARC. If your CryoSPARC installation uses the default base port of 39000, we can set up the required tunnels like so:
ssh -N -L 39000:localhost:39000 -L 39002:localhost:39002 -L 39003:localhost:39003 -L 39005:localhost:39005 << master host >>
Be sure to replace << master host >> (including the << and >>) with the hostname of the CryoSPARC master!
This command opens connections from port 39000 on your computer to port 39000 on the host, 39002 on your computer to 39002 on the host, etc. CryoSPARC tools needs these connections to function.
If your base port is not 39000, you’ll need to replace the ports as appropriate. For example, if your base port was 40000, you’d need to replace all instances of 39000 with 40000, 39002 with 40002, etc.
5. Create instance-info.json
Finally, you’ll need to provide your credentials to the cryosparc-tools scripts. These are your login credentials (typically your email and a password you chose), not any information about the CryoSPARC host or admin. Create a file called instance-info.json in your home directory and add the following information:
{
"license": "<< your license >>",
"email": "<< the email you use to log into the CryoSPARC GUI >>",
"password": "<< the password you use to log into the CryoSPARC GUI >>",
"base_port": << the base port used earlier (e.g., 39000)>>,
"host": "localhost"
}
replacing everything in and including << >> with the appropriate information. The license ID should be in the config.sh file you found earlier.
6. Run the script
Finally, you’re ready to run the scripts! If you create a python file (in this case, maybe get_particle_components.py) and add the script text to it, you can run it with python get_particle_components.py. Make sure you’re in the cs-tools conda environment!
For example, I’ll create the python file on my Desktop:
touch ~/Desktop/get_particle_components.py
and paste in the following text in the editor of my choice:
from cryosparc.tools import CryoSPARC
import json
import pandas as pd
from pathlib import Path
with open(Path.home() / 'instance-info.json', 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
# change this to whatever project and job you want to use
project_number = "P312"
job_number = "J54"
project = cs.find_project(project_number)
job = project.find_job(job_number)
particles = job.load_output("particles")
df = pd.DataFrame({
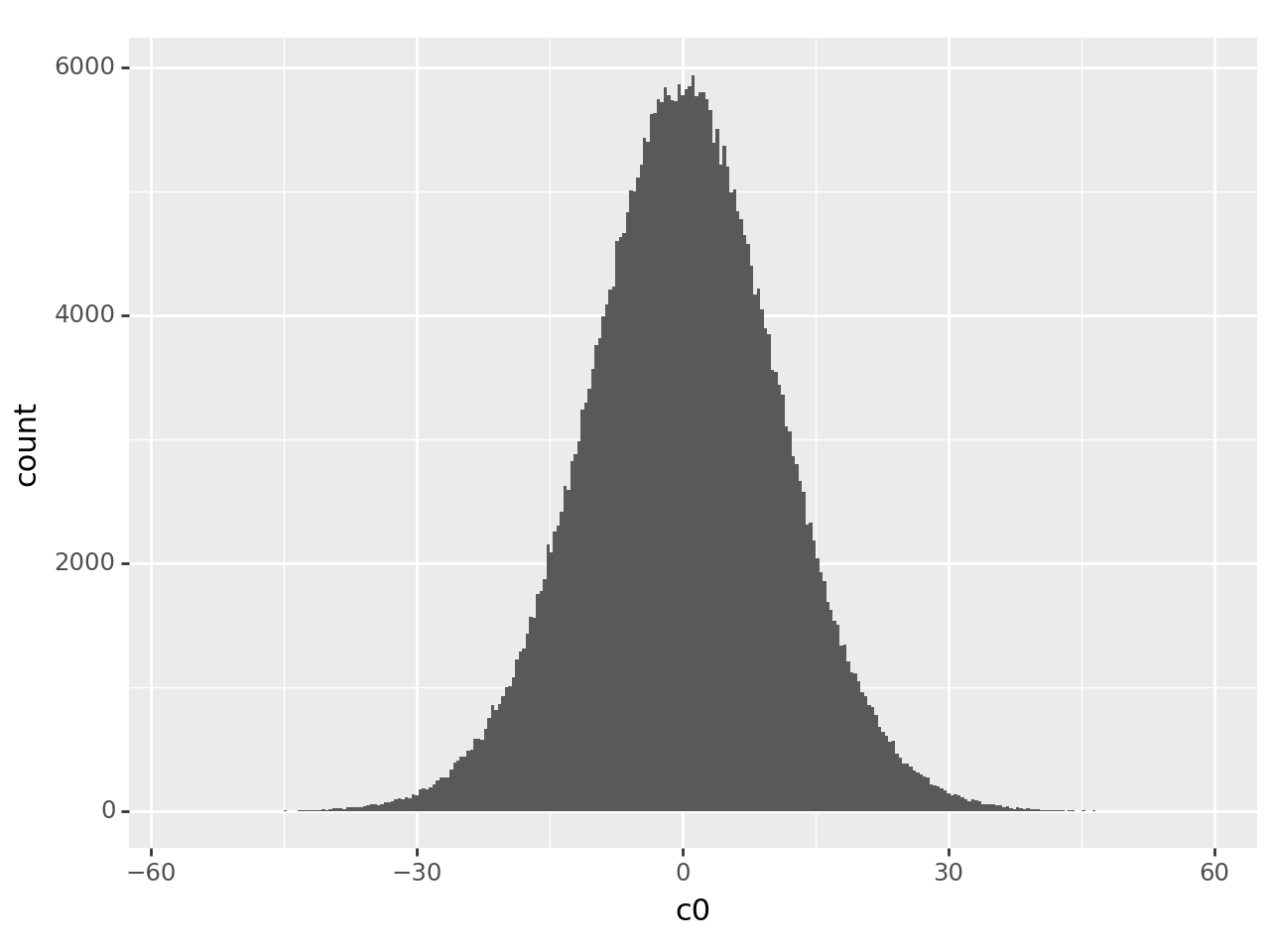
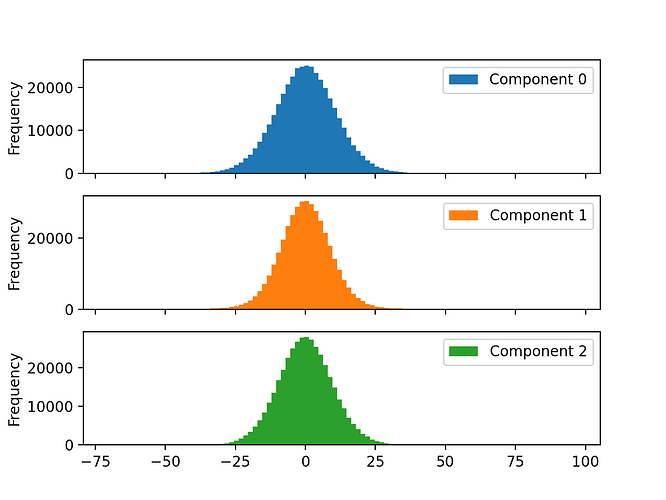
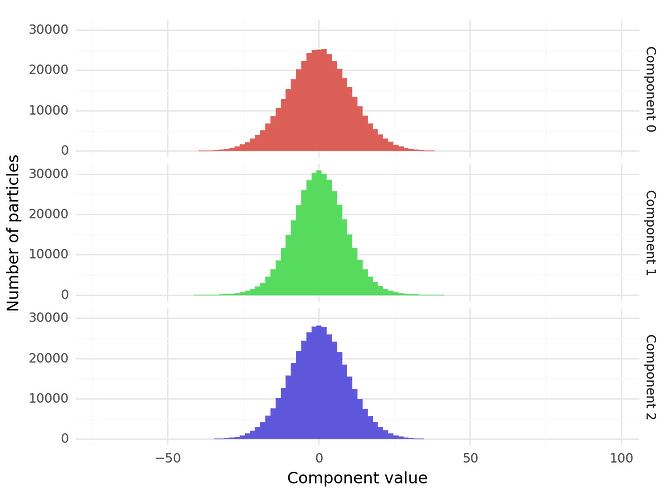
'c0': particles['components_mode_0/value'],
'c1': particles['components_mode_1/value'],
'c2': particles['components_mode_2/value']
})
print(df)
If I set up the SSH tunnels like in step 4, here is the result:
$ python ~/Desktop/get_particle_components.py
Connection succeeded to CryoSPARC command_core at http://localhost:39002
Connection succeeded to CryoSPARC command_vis at http://localhost:39003
Connection succeeded to CryoSPARC command_rtp at http://localhost:39005
c0 c1 c2
0 11.774458 13.445810 -20.135124
1 11.945024 2.822657 10.567183
2 -6.311073 0.321704 -17.275665
3 -0.410764 7.178520 0.903192
4 -41.858257 20.322187 -8.610754
... ... ... ...
397665 3.548247 4.151044 -12.174521
397666 0.581883 -13.682824 -10.587862
397667 0.757384 -11.042213 20.455528
397668 -2.539126 -5.445854 -1.673623
397669 7.511567 -0.164381 -3.030882
[397670 rows x 3 columns]
I hope that helps, and let me know if you run into any more problems!