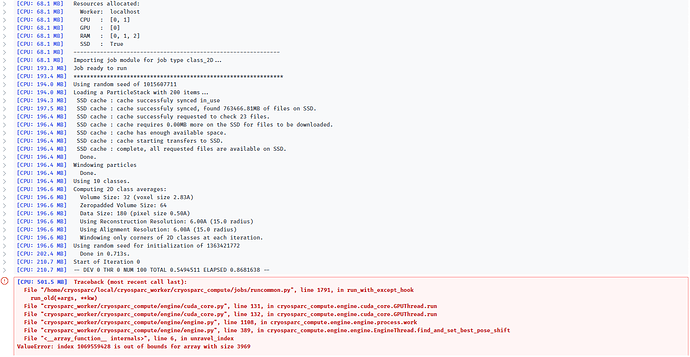
I manually picked 200 particles and tried to run a 2D classification job, but the job fails before even a single iteration is completed. All parameters are set to their default, other than the request to classify into 10 classes. Below is the output:
As similar index is out of bounds GPU-related errors have been reported in other posts, I will state that the workstation being used is running CentOS7. Below is the ouput of nvidia-smi:
I also checked the output of cryosparcm joblog for the failed job, which is not very informative:
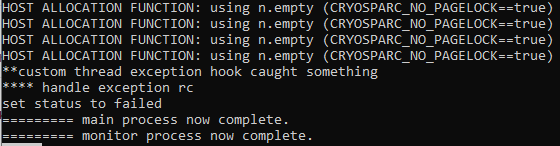
Two interesting observations:
-
In the same project where this failed job occurred, I ran two other 2D classification jobs using the output of blob picker (containing several hundred thousand particles). Both jobs completed successfully, although some runtime warnings were present in the
joblog. -
I tried to run this same 2D classification job multiple times, and the job does not always fail before the first iteration. In most cases it fails after iteration 1 with the above error, but in one instance it processed up to iteration 11 before failing with the above error.
Any advice on resolving this issue would be appreciated. Both Master and worker are running v3.2.0+210413.