Hello,
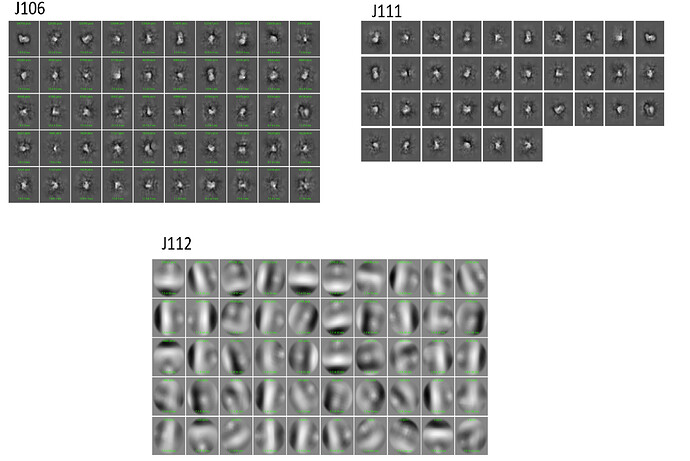
I am experiencing some issue with 2D classification of particles that were template-picked. The particles were picked, curated, then 2D classified using all default values (J106).
After weeding out the really bad classes, I was left with J111.
I attempted to do a second round of 2D classification. As the particles are small (~210 A on longest axis), I increased the number of iterations to 40 and the batch size to 400. These are the only parameters that I changed. The second round of 2D classification is shown in J112.
This is the first time I have had this issue and it only happens with template-picked particles. Within the past week, I did several rounds of 2D classification with blob-picked particles and never ran into this issue. Also, I have used cryosparc in the past with a template picker and not had this issue, so I am not sure what is going on.
Do you have any advice?
Did you try using a tighter circular mask?
The initial 2D classification did not use a circular mask, so I did not use one in the second iteration of 2D classes.
I went ahead and tried using a circular mask. The classes look better than J112, but still much worse than J106.
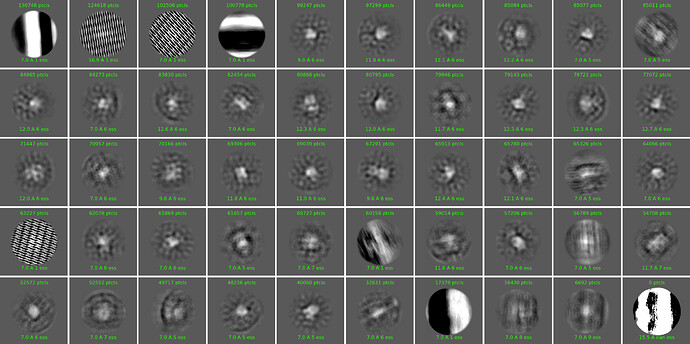
Looking at this image, J114, I have different advice. The weird classes in this image suggest to me that you have some bad micrographs. Probably there are low-mag atlas images that are getting particle picked by the template picker, but not the blob picker. I suggest going back and curating the micrographs. Alternatively, you could try eliminating all the bad classes from J114 and run the 2D class averages again, but I suspect you’re always going to have some of these bad particle from the bad micrographs, so curating the micrographs is a better option.
I find cryosparc’s micrograph curation a little tedious, and prefer to run an initial CTF correction in Relion. Then using Subset Selection in Relion, you can display all the power spectra at 0.25 size in a grid layout similar to how you view 2D class averages. Click to select all the bad ones, then right click and invert selection to select all the good ones and not the bad ones. I find it relatively easy to scan through 2,000-4,000 images and spot the ones with problematic power spectra. Finally, right click and save the subset, and import that curated micrograph subset into cryosparc.
YMMV
RJ