Hi,
When picking on some images in Live, I have observed an odd behavior:
- When using “Test Adjustments”, everything looks ok. The particles I want are picked from the test micrograph.
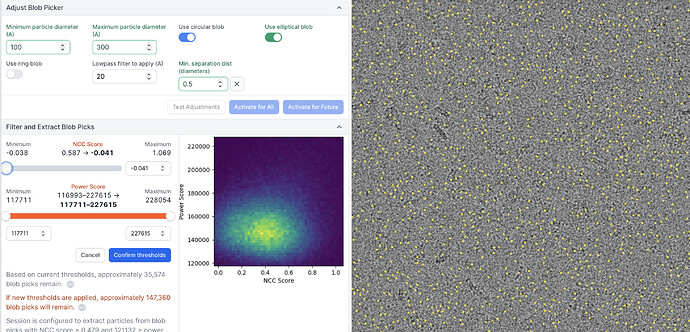
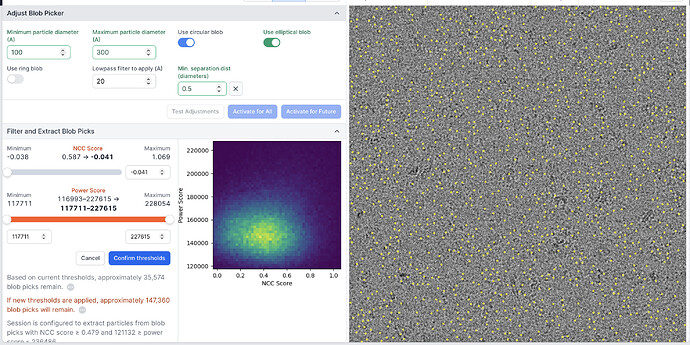
- When I click activate for all, and the micrograph is repicked as part of the full set, the picking changes - now what I consider to be the best, highest contrast particles are excluded - it seems to be picking everywhere but the particles that I want. See examples attached. This happens for both Template and blob picker, irrespective of parameters.
- If I attempt to now do “Test adjustments” again, it no longer picks the good particles in this mode either - it reproduces the behavior seen in step 2.
My theory as to what is going on is that it is applying some kind of power threshold, and excluding these particles based on contrast - as in the initial step, the “Test adjustments” step, these particles seemed to have the highest NCC values, at least using template picker. That is, the sliders do not contain the full contrast range of the micrograph. Is that possible, and if so how to fix?
Cheers
Oli
Hey @olibclarke,
This behaviour can be explained by how the histogram, NCC score and Power score are calculated.
First, the original power and NCC score ranges are calculated by the RTP worker. Then, the histogram is created by taking a random sample of 256 micrographs from the dataset, extracting the power and NCC scores of all the particles across all 256 (or less) micrographs, and taking the 1-99th percentile of values for each range.
I believe you were seeing better picks when in “Test” mode because the actual particle picks you’re looking for are probably in the top 1% of particles, which is captured when there are no thresholds placed on the picker. Once a threshold is set, the exposure viewer will only show particles within those thresholds.
Try this:
In cryosparc_master/command_vis/__init__.py, inside the function get_picker_hist(), edit the lines:
xvals = particles.data['pick_stats/ncc_score']
yvals = particles.data['pick_stats/power']
xmin = n.percentile(xvals, 0o1)
xmax = n.percentile(xvals, 99)
ymin = n.percentile(yvals, 0o1)
ymax = n.percentile(yvals, 99)
to
xvals = particles.data['pick_stats/ncc_score']
yvals = particles.data['pick_stats/power']
xmin = n.min(xvals)
xmax = n.max(xvals)
ymin = n.min(yvals)
ymax = n.max(yvals)
Save the file, then run cryosparcm stop command_vis && cryosparcm start command_vis
Then try checking if your picks show up when the thresholds are at their extremes.
2 Likes
Aha! - so in the event that one has very sparse particles, or a high percentage of bad micrographs (e.g. with gold etc) this is expected? Maybe it would be good to have a flag to turn this off (or change the threshold)? Maybe a disable thresholds checkbox, or something to show the full range of values? I can’t test this myself easily because it was on a core facility computer, but I will see if I can reproduce on my own setup