Welcome to the forum @guodong .
Please can you post
- post text instead of the screenshot.
You can obtain the text of the (end of the) event log with the commandcryosparcm eventlog P1 J590 | tail -n 50 - the outputs of these commands
cryosparcm cli "get_job('P1', 'J590', 'version', 'type', 'params_spec', 'started_at')" cryosparcm cli "get_job('P1', 'J513', 'version', 'type', 'params_spec', 'started_at')" cryosparcm eventlog P1 J513 | grep "Starting Topaz"
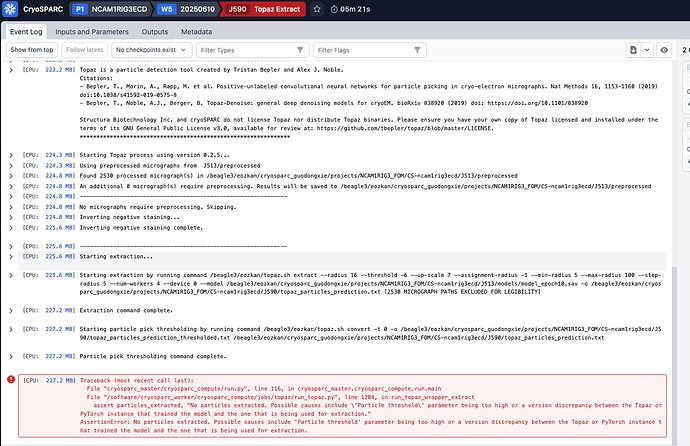
n 2025 16:14:44 GMT] [CPU RAM used: 224 MB] Starting Topaz process using version 0.2.5…
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 224 MB] Using preprocessed micrographs from J513/preprocessed
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] Found 2530 processed micrograph(s) in /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J513/preprocessed
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] An additional 0 micrograph(s) require preprocessing. Results will be saved to /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J513/preprocessed
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] --------------------------------------------------------------
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] No micrographs require preprocessing. Skipping.
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] Inverting negative staining…
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] Inverting negative staining complete.
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] --------------------------------------------------------------
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] Starting extraction…
[Wed, 18 Jun 2025 16:14:44 GMT] [CPU RAM used: 225 MB] Starting extraction by running command /beagle3/eozkan/topaz.sh extract --radius 16 --threshold -6 --up-scale 7 --assignment-radius -1 --min-radius 5 --max-radius 100 --step-radius 5 --num-workers 4 --device 0 --model /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J513/models/model_epoch10.sav -o /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J590/topaz_particles_prediction.txt [2530 MICROGRAPH PATHS EXCLUDED FOR LEGIBILITY]
[Wed, 18 Jun 2025 16:19:31 GMT] [CPU RAM used: 226 MB] Extraction command complete.
[Wed, 18 Jun 2025 16:19:31 GMT] [CPU RAM used: 226 MB] Starting particle pick thresholding by running command /beagle3/eozkan/topaz.sh convert -t 0 -o /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J590/topaz_particles_prediction_thresholded.txt /beagle3/eozkan/cryosparc_guodongxie/projects/NCAM1RIG3_FOM/CS-ncam1rig3ecd/J590/topaz_particles_prediction.txt
[Wed, 18 Jun 2025 16:19:35 GMT] [CPU RAM used: 226 MB] Particle pick thresholding command complete.
[Wed, 18 Jun 2025 16:19:35 GMT] [CPU RAM used: 226 MB] Traceback (most recent call last):
File “cryosparc_master/cryosparc_compute/run.py”, line 116, in cryosparc_master.cryosparc_compute.run.main
File “/software/cryosparc_worker/cryosparc_compute/jobs/topaz/run_topaz.py”, line 1204, in run_topaz_wrapper_extract
assert particles_extracted, “No particles extracted. Possible causes include 'Particle threshold' parameter being too high or a version discrepancy between the Topaz or PyTorch instance that trained the model and the one that is being used for extraction.”
AssertionError: No particles extracted. Possible causes include ‘Particle threshold’ parameter being too high or a version discrepancy between the Topaz or PyTorch instance that trained the model and the one that is being used for extraction.
cryosparcm cli “get_job(‘P1’, ‘J590’, ‘version’, ‘type’, ‘params_spec’, ‘started_at’)”
{‘_id’: ‘685070bbb9b8e5d067102141’, ‘params_spec’: {‘exec_path’: {‘value’: ‘/beagle3/eozkan/topaz.sh’}, ‘par_diam’: {‘value’: 150}}, ‘project_uid’: ‘P1’, ‘started_at’: ‘Wed, 18 Jun 2025 16:14:39 GMT’, ‘type’: ‘topaz_extract’, ‘uid’: ‘J590’, ‘version’: ‘v4.6.0’}
A post was merged into an existing topic: Hetero refinement issue
Thanks for posting the information for job J590. Please can you also post information for the ancestor job J513, which you can obtain with the commands
cryosparcm cli "get_job('P1', 'J513', 'version', 'type', 'params_spec', 'started_at')"
cryosparcm eventlog P1 J513 | grep "Starting Topaz"