Hi CS user,
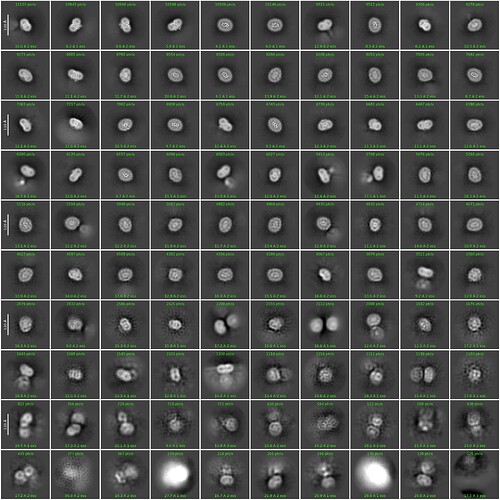
I have a 70 kd membrane protein where I have very nice 2D classes (attached) with visible secondary feature.
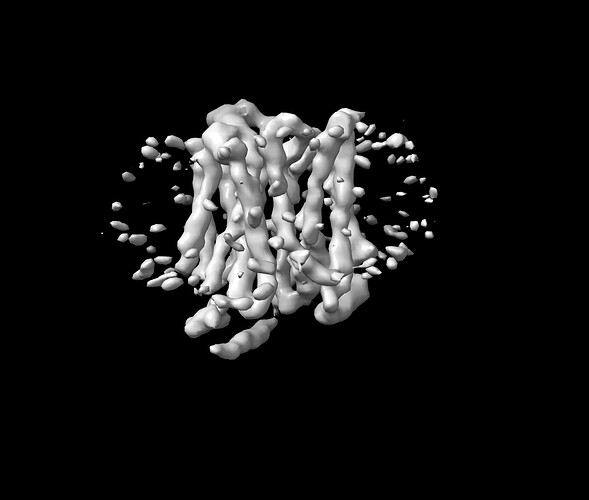
My NU-refinement job provide me a map with 12 transmembrane domain visible but the map is not good quality.
Any trick / suggestion I should try to improve it?
Best
Nitesh
`

`
.
2 Likes
Very nice data! Some info on refinement settings and what you have tried (in terms of 3D classification etc) might be helpful so folks can suggest how to improve 
1 Like
Hi Oli,
I have made 3 ab-initio classes in CS with 20K particles form the 2D classes. I used them as reference map and performed heterogeneous refinement on all particles from 2D class.
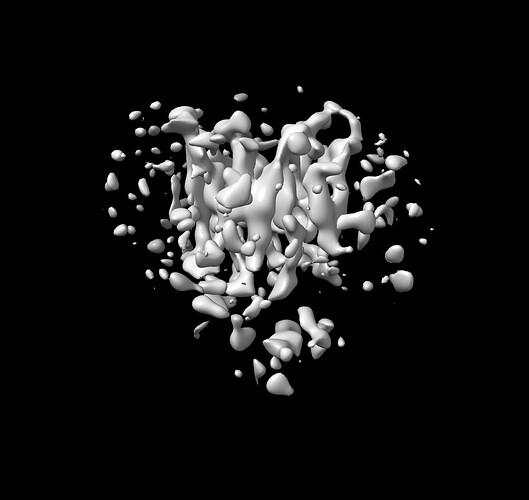
Out of the three classes I picked the one best and performed NU (attached image).
NU refinement setting:
I used Box size 440
number of extra final pass:2
max alignment res: 7
Initial low pass filter :20
Further I remove micelle around it and performed the local refinement (attached).
I can see 12 TMs in my map but no distinguish secondary features.
Best
Nitesh
1 Like
What settings did you use for ab initio?
Number of Class 3
particle used 20K
Max resolution:7
Initial resolution:20
Initial minibatch size :300
Final minibatch size 1500
1 Like
Did you see clear helices in ab initio? Sometimes for small membrane proteins without much protruding from the micelle, using higher res info in ab initio can work better (and not restricting the number of particles). E.g. I would try an initial res of 9 and final res of 7, or even start res of 7 and final res of 5.
2 Likes
I didn’t see helical in ab-initio but some density inside the micelle.
I will try your suggestion. With roughly 400K particles should I go for 4 ab-initio class rather than 3?
I mean is there a particle vs class ratio I should considered?
Thank you.
No, three is fine - sometimes I would even start out with a single-class ab initio (faster) if the particle set is fairly clean
I would expect based on your 2Ds that you ought to see clear helical density in ab initio (as it is clearly evident from your 2D classes)
Thank you Oli for the suggestions.
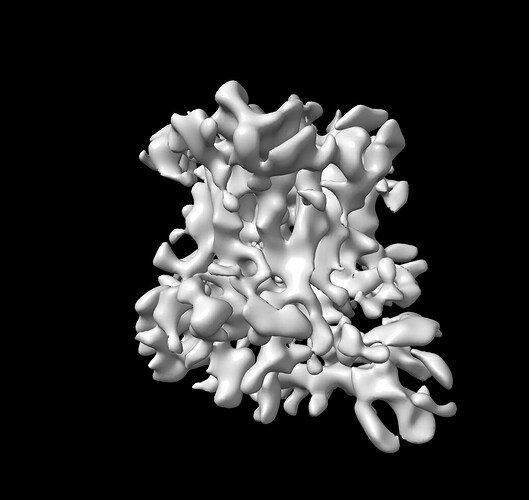
I applied your trick and my ab-initio map improved (attached image).
I will start the NU on this map.
Best
Nitesh
4 Likes
ah looks lovely that’s great!! good luck!
1 Like
Hi, I have also been making proteins with similar molecular weight recently. I would like to ask you what the photos of your protein particles look like? Is it convenient to have a look, thank you.
Hi Linqian,
The particle were pretty visible in my case. I can see round particle density contributed by particle with detergent micelle.
As you can see my 2D classification also give very good results (around 400K particles).
1 Like