Hello everyone,
I am using V4.2 currently and I have encountered a strange error when processing my data.
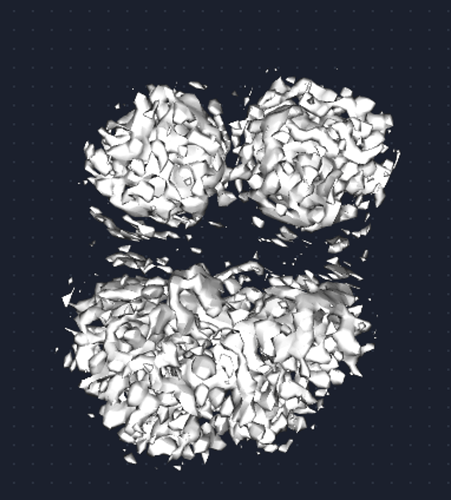
I am working with a small membrane protein that is ~20 kDa as a monomer… Attached to each monomer is a GFP to act as a fiducial. The protein forms a dimer, which makes the total mass of the complex ~100 kDa.
The issue that I am having is that the micelle belt around the membrane protein remains, and the density where the protein should be disappears as shown below.
I have run particle subtraction to remove the GFP, but prior to subtraction the map looks like this, which indicates that the dimer is present within the micelle, but just not visible after refinement. Has anyone experienced this before?