Hi @rposert,
I ran into the same issue when using cryoSPARC tools in a Jupyter notebook. Could you help me diagnose what might be happening?
Initially, I noticed that the version number in the upper-left corner in the cryoSPARC home page showed “null”, and a response by wtempel in this thread suggested that restarting cryoSPARC would help. While this did fix the version number, it didn’t have any effect on the connection reset by peer error.
Based on your comment, I also checked to make sure my versions of cryoSPARC and cryosparc-tools match:

This is a little strange, because the notebook appears to be connecting successfully:
cs = CryoSPARC(
license="redacted",
host="localhost",
base_port=39000,
email=redacted",
password="redacted"
)
assert cs.test_connection()
Connection succeeded to CryoSPARC command_core at http://localhost:39002
Connection succeeded to CryoSPARC command_vis at http://localhost:39003
Connection succeeded to CryoSPARC command_rtp at http://localhost:39005
Moreover, I can successfully import the results of a job into the notebook. The error only occurs when I try to save a results group.
for cluster_id, inds in clusters.items():
cluster_cs = J380_particles.take(inds)
cluster_cs_name = f'cluster_{cluster_id}/{num_clusters}'
print(f'{cluster_cs_name}: len({cluster_cs.rows()}) particles')
print(cluster_cs_name)
cs.save_external_result('P10',
'W17',
cluster_cs,
type="particle",
name=cluster_cs_name,
slots=["blob"],
passthrough=('J380', "particles"),
title=cluster_cs_name
)
cluster_0/10: len(Spool object with 149935 items.) particles
cluster_0/10
/programs/x86_64-linux/anaconda/2022.10/envs/cs-tools/lib/python3.8/contextlib.py:113: UserWarning: *** CommandClient: (http://localhost:39003/external/projects/P10/jobs/J700/outputs/cluster_0/10/dataset) URL Error [Errno 104] Connection reset by peer, attempt 1 of 3. Retrying in 30 seconds
return next(self.gen)
/programs/x86_64-linux/anaconda/2022.10/envs/cs-tools/lib/python3.8/contextlib.py:113: UserWarning: *** CommandClient: (http://localhost:39003/external/projects/P10/jobs/J700/outputs/cluster_0/10/dataset) URL Error [Errno 104] Connection reset by peer, attempt 2 of 3. Retrying in 30 seconds
return next(self.gen)
Inside the project, a results group is generated. But although the job appears to have stopped with an error, there isn’t an obvious error message in the log:
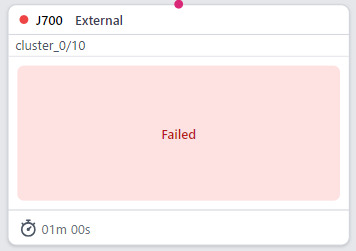
[CPU: 268.3 MB Avail: 184.32 GB]
Adding input slot particles (type particle)...
[CPU: 268.3 MB Avail: 184.31 GB]
Created input group: particles (type particle)
[CPU: 268.3 MB Avail: 184.31 GB]
Adding output group cluster_0/10 (type particle)...
[CPU: 268.3 MB Avail: 184.31 GB]
Created output group cluster_0/10 (type particle, passthrough particles)
[CPU: 268.3 MB Avail: 184.32 GB]
Added output slot for group cluster_0/10: blob (type particle.blob)
[CPU: 268.3 MB Avail: 184.32 GB]
Passed through from input particles to output cluster_0/10
[CPU: 268.3 MB Avail: 184.32 GB]
Passed through J380.particles to J700.particles
License is valid.
Any insights would be greatly appreciated!
Best,
cbeck

