Hi,
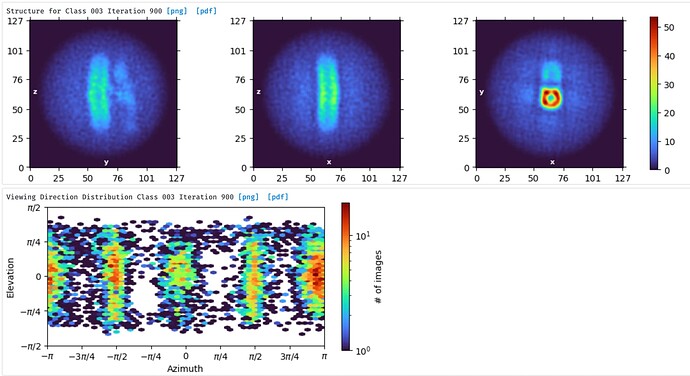
In cases where we have all side views, and no top views, of a disk-shaped sample, ab initio frequently fails in this mode, where we end up with a cylinder, and all the different side views are assigned to views orthogonal to the long axis of the cylinder.
Is there any way to introduce some kind of “shape” prior to prevent this? E.g. ab-initio, but instead of starting from random density, start from a disk, or similar?
In a case like this the underlying data is fine, and can give a high resolution reconstruction if refined against the correct model, it is just ab initio that is failing.
There is also another, related failure mode we have observed, where we have side views and “oblique” views, but no top views, whereupon the oblique views are assigned as top views. That is a little more dangerous, as it almost looks sensible (just a little squashed), and will give decent resolution (internally self consistent) but incorrect reconstructions upon refinement.
Cheers
Oli