Hi all,
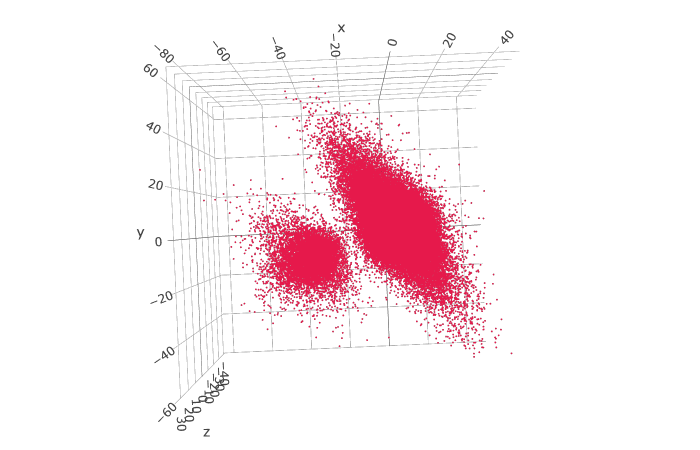
I’m working on a protein which contains both discrete and continuous heterogeneity which can be captured by 3DVA, as seen here:
There are two obvious clusters of particles here, and I’d like to be able to sort/separate the particles by which cluster they belong to, and then use the sorted particles in each cluster to perform a consensus refinement (this would be somewhat analogous to the particle filtering function in cryoDRGN). I was wondering if there was an easy way to do this on cryoSPARC? Also, just in general, is it possible to separate the particles directly by the volume they contributed to generating along a trajectory in the 3DVA job (such that a further consensus reconstruction could be performed on the particles that went into each intermediate volume)? Or, is this further refinement not worthwhile because 3DVA has already done the best alignments/refinements it can against each individual intermediate volume along the way, and redoing refinement on a subset of the particles would not yield any higher resolution map? Lastly, is there a way to visualize where along the plot of the components each individual map came from? Any advice on whether the above are achievable (and if not, if there is a way to mimic the aforementioned functionality in cryoSPARC) would be greatly appreciated.
Many thanks in advance for everyone’s input!
Best,
Adele