Hi all,
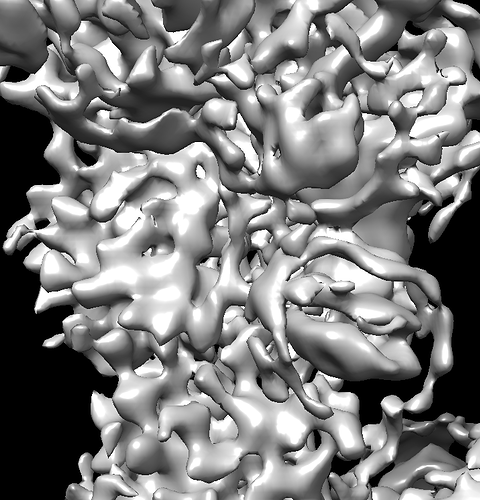
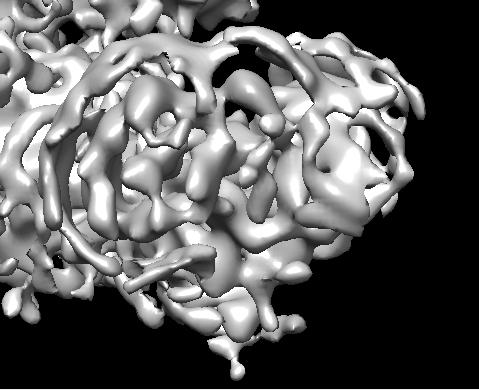
I’m working on a protein with an approximate molecular weight of 150 kDa. Despite collecting a large dataset, the EM map resolution appears to be stuck at around 6 Å.
One of the main issues I’m encountering is the appearance of ring-like artifacts around the map, and it also seems to be over-sharpened.
Could anyone suggest how to minimize or avoid these artifacts during map reconstruction or post-processing?
I’m attaching a few EM map snapshots for your reference.
Thanks in advance for your support!