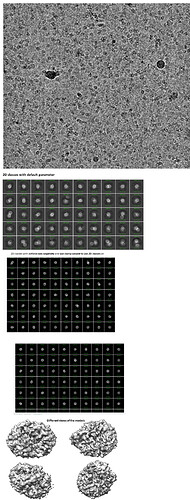
Hi all,
I collected a data set on Krios for a small membrane-anchored protein (40kDa) with three transmembrane helices. I purified this protein in 0.05% DDM and the size exclusion chromatography shows a single peak of an oligomer (not sure about the oligomeric state of the protein, could be dimer/tetramer). I could see decent particles in the micrograph. When I did 2D classification with the default settings, I saw blurry classes. When I turned the force max off with 40 EM iterations and batch size: 400 along with enforcing non-negativity and clamp solvent, I could see some features in the 2D classes. I built an ab initio model with a start resolution: 9 A and a final resolution of 5 A. The shape of the ab initio model seems to be different from the model I got from the Alpha fold. Please guide me on how to confirm if I am looking at detergent micelles only or protein embedded in detergent micelles.
Thanks!
Hi,
It’s a tricky project for sure, as you have a small protein, plus a small detergent belt. With 3 TM, you might expect say 100-150 DDM forming the belt, and out of these, maybe half would be visible in reconstructions, and share enough positions to count in the alignments/reconstructions.
You only show 1 ab-initio, did you try several? With your 2D classes, it seems like there are definitely DDM micelles in them, but maybe(?) other stuffs. So your challenge would be to distinguish the two, and it might work by asking more ab-initio models, and/or refining your 2D classes.
Good luck