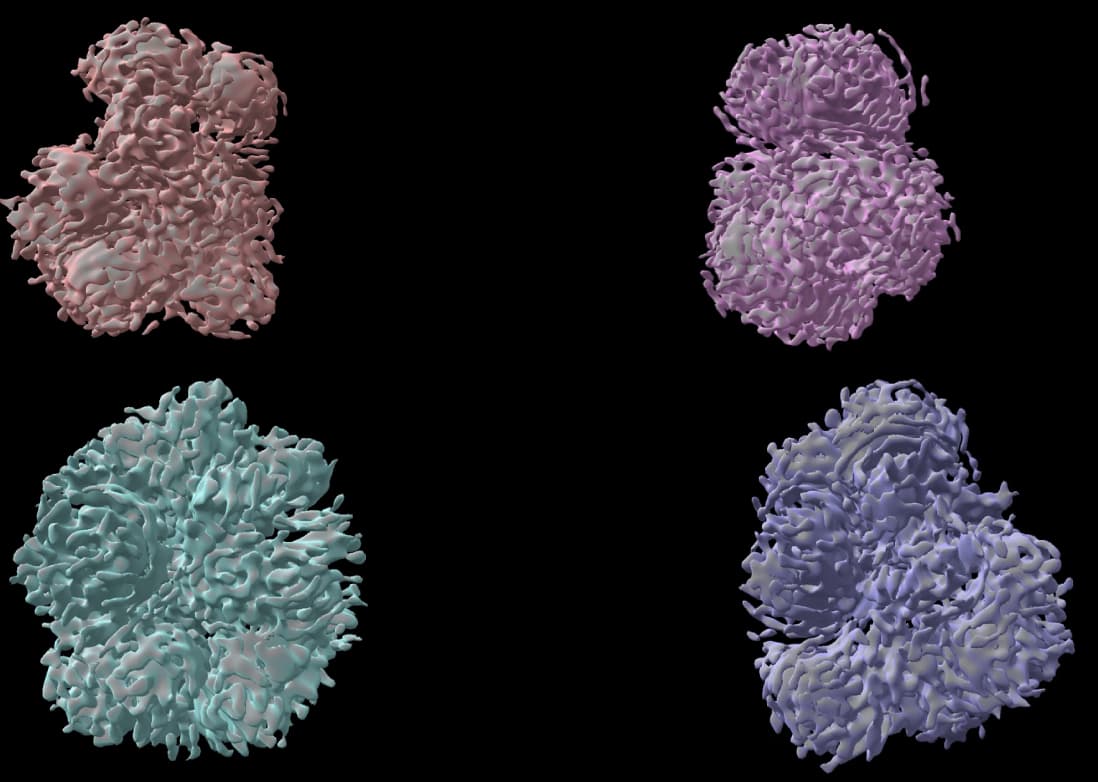
Hello everyone, my protein is a polymer, which is polymerized by more than a dozen proteins, and I have obtained several fixed structures through several iterations such as three-dimensional classification, template selection particles, can I consider them all real? In the follow-up, I made a mask for local refinement, but the effect was not particularly good, so for this nonsymmetrical protein polymer structure analysis, do you have any good suggestions for me, thank you very much!
Hi Heiqixin,
It is very hard to offer advice without more information - without seeing data (example classes etc) or more detailed descriptions of what you are having trouble with, it is difficult to draw any conclusions.
Cheers
Oli
I checked the relevant literature, I didn’t find any relevant reports on cryo-EM analysis of polymers, these are the four structures I got, the protein I want to parse has not been studied before, I express it as a polymer, I don’t know what to do next now, cryosprc shows that its resolution rate is about 6, do you think its resolution is still possible to improve? Or would the mask help? Thank you very much!
Hi,
Can you show these volumes at higher threshold? And/or perhaps screenshots from the refinement logs?
If your resolution is 6Å, you should start to see secondary structure (helices) at higher thresholds.
Also, how does the overall molecular envelope compare to what you would expect from structure prediction algorithms (Alphafold etc)
Cheers
Oli
I realise that this point is just being pedantic, but all proteins are polymers. ![]()
What do the 2D classes look like? Would you show them? And as @olibclarke requests, the 3D at a higher threshold? Those density maps look purely overfitted, and I can’t see any sort of expected secondary structure (again, as previously stated, helices should be pretty obviously defined at 6Å) hiding under the artefacts. What does the angular distribution look like?
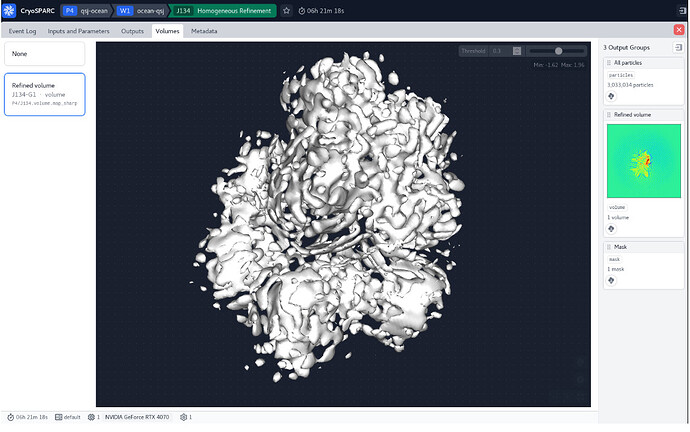
Thank you for your professional answers! When I raise the threshold, the structure becomes very empty. Nor is there a clear secondary structure in sight. It looks like a bud and doesn’t look like a homologous modeled structure.
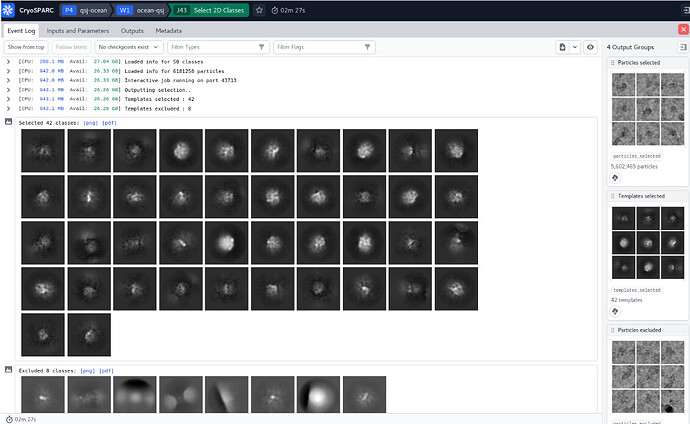
Thank you for your professional answers! My 2D structure doesn’t seem to be very clear, do you think there is a need for further optimization of my batch of data?
The 3D is overfitted noise, and there is little in the way of clarity in the 2D. You can try increasing the number of classes (try 200) and iterations (try 30) for the 2D to see if you can find anything (as the particles on the right definitely look like there is something there)… also try setting the initial classification uncertainty factor to 5 or 8 to slow class convergence…
Thank you very much for your help. I modified the 2D parameters according to your suggestions, and the 2D classification I got still seems not optimistic as shown in the figure. May I ask if my batch of data is not good?
Going a step back, do you clearly see the particles in your micrographs? Is your picking mostly picking those particles, or are there lots of false positives? Perhaps you could share images of a few micrographs, at low and high defocus.
A micrograph showing picks would be helpful too. Cut down the number of picks (increase NCC threshold) to try removing picks which have less signal and are more likely to be noise rather than particles.