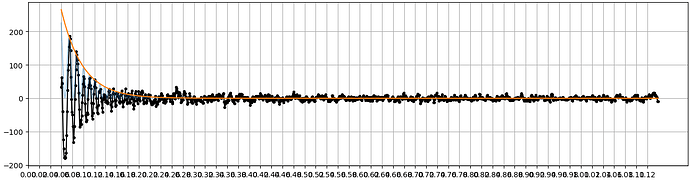
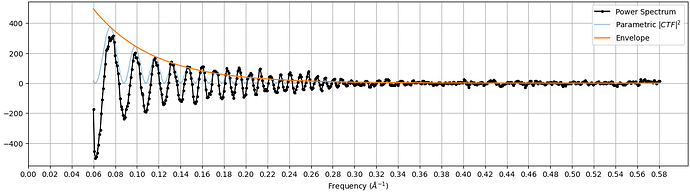
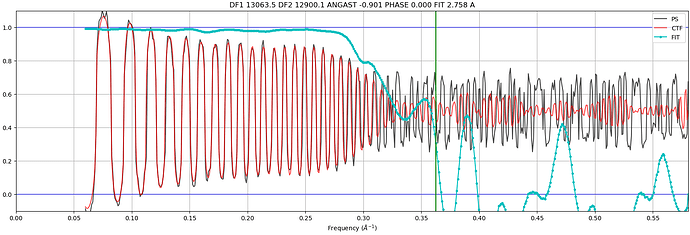
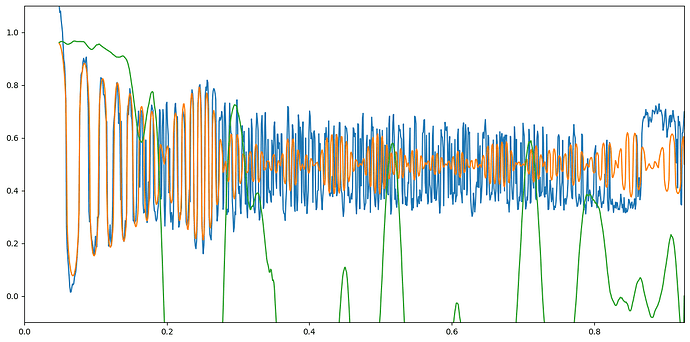
Hi @subhrob15! If you’re asking about the plot with the power spectrum, CTF, and cross correlation you can indeed access that data. In the directory of the Patch CTF job, you’ll see a directory called ctfestimated. Within that directory are a number of .npy files (one for each micrograph), that contain the data transformed to produce these plots.
For instance, I have a Patch CTF job J49. I can get a list of all the relevant files like so:
import numpy as np
from pathlib import Path
ctf_job_path = Path("/bulk1/data/cryosparc_personal/rposert/CS-rposert-guide-work/J49")
all_ctf_files = (ctf_job_path / "ctfestimated").glob('*diag_plt.npy')
Then, if I write a function to plot them:
import matplotlib.pyplot as plt
def make_ctf_plot(data, filename):
# note that raw data must be transformed
power_spectrum = np.cbrt(data['EPA_trim'] - data['BGINT']) + 0.5
ctf = np.cbrt(data['ENVINT'] * (2 * data['CTF']**2 - 1)) + 0.5
cc = data['CC']
# make plot
plt.figure(figsize = [12, 6])
plt.plot(data['freqs_trim'], power_spectrum)
plt.plot(data['freqs_trim'], ctf)
plt.plot(data['freqs_trim'], cc)
plt.ylim([-0.1, 1.1])
plt.xlim(0, data['freqs_trim'].max())
plt.tight_layout()
plt.savefig(filename)
and/or save to CSV for use in other software:
import pandas as pd
def write_csv(data, filename):
df = pd.DataFrame({
'freq': data['freqs_trim'],
'ps': np.cbrt(data['EPA_trim'] - data['BGINT']) + 0.5,
'ctf': np.cbrt(data['ENVINT'] * (2 * data['CTF']**2 - 1)) + 0.5,
'cc': data['CC']
})
df.to_csv(filename, index = False)
I can write out the data for each micrograph like so:
for ctf_file in all_ctf_files:
data = np.load(ctf_file)
plot_name = ctf_file.name.replace('.npy', '.png')
make_ctf_plot(data, plot_name)
csv_name = ctf_file.name.replace('.npy', '.csv')
write_csv(data, ctf_file.name.replace('.np', '.csv'))
Note that this script would create a plot and CSV for every single one of your micrographs — you probably do not want to generate that many files!
If you were referring to the other plot (with Power Spectrum, Parametric |CTF|^2, and Envelope), that data is not available outside the job.
I hope that helps!