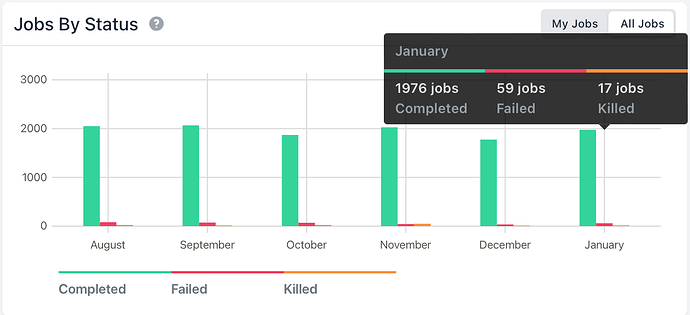
Here is the command core log:
> 2024-01-24 11:08:56,468 scheduler_run_core INFO | Jobs Queued: [('P386', 'J79'), ('P31', 'J3')]
> 2024-01-24 11:08:56,472 scheduler_run_core INFO | Licenses currently active : 8
> 2024-01-24 11:08:56,472 scheduler_run_core INFO | Now trying to schedule J79
> 2024-01-24 11:08:56,472 scheduler_run_job INFO | Scheduling job to CBE
> 2024-01-24 11:08:57,710 scheduler_run_job INFO | Not a commercial instance - heartbeat set to 12 hours.
> 2024-01-24 11:08:57,780 scheduler_run_job INFO | Launchable! -- Launching.
> 2024-01-24 11:08:57,787 set_job_status INFO | Status changed for P386.J79 from queued to launched
> 2024-01-24 11:08:57,788 app_stats_refresh INFO | Calling app stats refresh url http://imp-cryosparc-1.vbc.ac.at:39000/api/actions/stats/refresh_job for project_uid P386, workspace_uid None, job_uid J79 with body {'projectUid': 'P386', 'jobUid': 'J79'}
> 2024-01-24 11:08:57,794 app_stats_refresh INFO | code 200, text {"success":true}
> 2024-01-24 11:08:57,820 run_job INFO | Running P386 J79
> 2024-01-24 11:08:57,844 run_job INFO | cmd: ssh cbe.vbc.ac.at sbatch --wckey=cryosparc /groups/haselbach/cryosparc_projects_folder/active_instance/imports/CS-231229-helmutatpys/J79/queue_sub_script.sh
> 2024-01-24 11:08:59,066 scheduler_run_core INFO | Licenses currently active : 9
> 2024-01-24 11:08:59,066 scheduler_run_core INFO | Now trying to schedule J3
> 2024-01-24 11:08:59,067 scheduler_run_core INFO | Queue status waiting_licenses
> 2024-01-24 11:08:59,067 scheduler_run_core INFO | Queue message
> 2024-01-24 11:08:59,068 scheduler_run_core INFO | Finished
> 2024-01-24 11:08:59,092 scheduler_run_core INFO | Running...
> 2024-01-24 11:08:59,092 scheduler_run_core INFO | Jobs Queued: [('P31', 'J3')]
> 2024-01-24 11:08:59,095 scheduler_run_core INFO | Licenses currently active : 9
> 2024-01-24 11:08:59,095 scheduler_run_core INFO | Now trying to schedule J3
> 2024-01-24 11:08:59,095 scheduler_run_core INFO | Queue status waiting_licenses
> 2024-01-24 11:08:59,095 scheduler_run_core INFO | Queue message
> 2024-01-24 11:08:59,096 scheduler_run_core INFO | Finished
> ESC[0mSending command: ssh cbe.vbc.ac.at squeue -j 65347214 ESC[0m
> ESC[0mSending command: ssh cbe.vbc.ac.at squeue -j 64918082 ESC[0m
> ESC[0mCluster job status check failed with exit code 1ESC[0m
> ESC[0mslurm_load_jobs error: Invalid job id specified
> ESC[0m
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | Cluster job status update for P24 J261 failed with exit code 1
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | Traceback (most recent call last):
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_command/command_core/__init__.py", line 2883, in update_cluster_job_status
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | cluster_job_status = cluster.get_cluster_job_status(target, cluster_job_id, template_args)
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_compute/cluster.py", line 176, in get_cluster_job_status
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | res = subprocess.check_output(shlex.split(cmd), stderr=subprocess.STDOUT).decode()
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 415, in check_output
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | return run(*popenargs, stdout=PIPE, timeout=timeout, check=True,
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 516, in run
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | raise CalledProcessError(retcode, process.args,
> 2024-01-24 11:09:02,640 update_cluster_job_status ERROR | subprocess.CalledProcessError: Command '['ssh', 'cbe.vbc.ac.at', 'squeue', '-j', '64918082']' returned non-zero exit status 1.
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | slurm_load_jobs error: Invalid job id specified
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | Traceback (most recent call last):
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_command/command_core/__init__.py", line 2883, in update_cluster_job_status
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | cluster_job_status = cluster.get_cluster_job_status(target, cluster_job_id, template_args)
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_compute/cluster.py", line 176, in get_cluster_job_status
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | res = subprocess.check_output(shlex.split(cmd), stderr=subprocess.STDOUT).decode()
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 415, in check_output
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | return run(*popenargs, stdout=PIPE, timeout=timeout, check=True,
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 516, in run
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | raise CalledProcessError(retcode, process.args,
> 2024-01-24 11:09:02,642 update_cluster_job_status ERROR | subprocess.CalledProcessError: Command '['ssh', 'cbe.vbc.ac.at', 'squeue', '-j', '64918082']' returned non-zero exit status 1.
> 2024-01-24 11:09:04,473 dump_workspaces INFO | Exporting all workspaces in P197...
> 2024-01-24 11:09:04,495 dump_workspaces INFO | Exported all workspaces in P197 to /groups/haselbach/cryosparc_projects_folder/active_instance/imports/20221030_Doa10_BRIL_Nb_nanodisc_b3_g1/workspaces.json in 0.02s
> 2024-01-24 11:09:04,677 scheduler_run_core INFO | Running...
> 2024-01-24 11:09:04,678 scheduler_run_core INFO | Jobs Queued: [('P31', 'J3')]
> 2024-01-24 11:09:04,681 scheduler_run_core INFO | Licenses currently active : 9
> 2024-01-24 11:09:04,681 scheduler_run_core INFO | Now trying to schedule J3
> 2024-01-24 11:09:04,681 scheduler_run_core INFO | Queue status waiting_licenses
> 2024-01-24 11:09:04,681 scheduler_run_core INFO | Queue message
> 2024-01-24 11:09:04,682 scheduler_run_core INFO | Finished
> 2024-01-24 11:09:06,256 set_job_status INFO | Status changed for P386.J79 from launched to started
> 2024-01-24 11:09:06,258 app_stats_refresh INFO | Calling app stats refresh url http://imp-cryosparc-1.vbc.ac.at:39000/api/actions/stats/refresh_job for project_uid P386, workspace_uid None, job_uid J79 with body {'projectUid': 'P386', 'jobUid': 'J79'}
> 2024-01-24 11:09:06,265 app_stats_refresh INFO | code 200, text {"success":true}
> 2024-01-24 11:09:07,356 set_job_status INFO | Status changed for P386.J79 from started to running
> 2024-01-24 11:09:07,358 app_stats_refresh INFO | Calling app stats refresh url http://imp-cryosparc-1.vbc.ac.at:39000/api/actions/stats/refresh_job for project_uid P386, workspace_uid None, job_uid J79 with body {'projectUid': 'P386', 'jobUid': 'J79'}
> 2024-01-24 11:09:07,367 app_stats_refresh INFO | code 200, text {"success":true}
> 2024-01-24 11:09:09,737 scheduler_run_core INFO | Running...
> 2024-01-24 11:09:09,737 scheduler_run_core INFO | Jobs Queued: [('P31', 'J3')]
> 2024-01-24 11:09:09,742 scheduler_run_core INFO | Licenses currently active : 9
> 2024-01-24 11:09:09,743 scheduler_run_core INFO | Now trying to schedule J3
> 2024-01-24 11:09:09,743 scheduler_run_core INFO | Queue status waiting_licenses
> 2024-01-24 11:09:09,743 scheduler_run_core INFO | Queue message
> 2024-01-24 11:09:09,745 scheduler_run_core INFO | Finished
> ESC[0mSending command: ssh cbe.vbc.ac.at squeue -j 64918082 ESC[0m
> ESC[0mCluster job status check failed with exit code 1ESC[0m
> ESC[0mslurm_load_jobs error: Invalid job id specified
> ESC[0m
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | Cluster job status update for P24 J261 failed with exit code 1
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | Traceback (most recent call last):
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_command/command_core/__init__.py", line 2883, in update_cluster_job_status
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | cluster_job_status = cluster.get_cluster_job_status(target, cluster_job_id, template_args)
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_compute/cluster.py", line 176, in get_cluster_job_status
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | res = subprocess.check_output(shlex.split(cmd), stderr=subprocess.STDOUT).decode()
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 415, in check_output
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | return run(*popenargs, stdout=PIPE, timeout=timeout, check=True,
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 516, in run
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | raise CalledProcessError(retcode, process.args,
> 2024-01-24 11:09:13,656 update_cluster_job_status ERROR | subprocess.CalledProcessError: Command '['ssh', 'cbe.vbc.ac.at', 'squeue', '-j', '64918082']' returned non-zero exit status 1.
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | slurm_load_jobs error: Invalid job id specified
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | Traceback (most recent call last):
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_command/command_core/__init__.py", line 2883, in update_cluster_job_status
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | cluster_job_status = cluster.get_cluster_job_status(target, cluster_job_id, template_args)
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/cryosparc_compute/cluster.py", line 176, in get_cluster_job_status
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | res = subprocess.check_output(shlex.split(cmd), stderr=subprocess.STDOUT).decode()
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 415, in check_output
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | return run(*popenargs, stdout=PIPE, timeout=timeout, check=True,
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | File "/home/svc_cryosparc/cryosparc2_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.8/subprocess.py", line 516, in run
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | raise CalledProcessError(retcode, process.args,
> 2024-01-24 11:09:13,657 update_cluster_job_status ERROR | subprocess.CalledProcessError: Command '['ssh', 'cbe.vbc.ac.at', 'squeue', '-j', '64918082']' returned non-zero exit status 1.