Hello,
I am working on a complex of the 40S ribosomal subunit with different semi-flexible components attached. I have had good results disentangling flexibility with 3DVA clustering, but then when I try to refine the clusters furthers, the 40S dominates the alignments and my smaller protein gets blurry again.
Running local refinement helped a little bit, but I want to perform particle subtraction on most of the 40S to further improve the input particles for the local refinement.
I prepared a mask (using segger in chimera - which worked well to generate masks for the 3DVA), resampled on a correct volume, imported it, dilated by 5 and padded by 5 using the volume tools, but then the particle subtraction runs and the particles look the same as before.
I went through the forums and found the suggestion to run a local refinement with the subtraction mask before subtraction, which I did, but nothing changed.
I tried running a 2D classification and a 3D homogeneous refinement with the supposedly subtracted particles, and in both cases the results look the same as before the particle subtraction.
I went through the parameters of the particle subtraction job, but there is not really that much to change (to my inexperienced eyes).
We are running cryosparc 3.1.
Any suggestions?
lg
Hi @cryo-lg,
Thanks for reporting this. Could you let us know the box size of the volume, mask, and particles going into the particle subtraction job, as well as any modified parameters? As of now, particle subtraction requires that all sizes be equal for the scaling to be correct – this is potentially the issue if the box sizes differ (if this is the case, you may notice a warning in the job log saying WARNING: Resampling input volume - this is untested, the scale might be off – unfortunately this isn’t printed to the streamlog though, so it can fly by unnoticed).
Also, are you able to share the plots from the particle subtraction job showing the FSC, the “Lowpass filtered, subtracted images”, and the “Lowpass filtered, original images”? (If you prefer, you can share this with me over DM).
Thanks,
Michael
Hello,
of course: box size of everything is 512. I thought this was a requirement anyway.
I am not seeing any warning.
[CPU: 2.09 GB] --------------------------------------------------------------
[CPU: 2.09 GB] Compiling job outputs…
[CPU: 2.09 GB] Passing through outputs for output group particles from input group particles
[CPU: 2.09 GB] This job outputted results [‘blob’]
[CPU: 2.09 GB] Loaded output dset with 27428 items
[CPU: 2.09 GB] Passthrough results [‘ctf’, ‘alignments3D’, ‘components_mode_0’, ‘components_mode_1’, ‘components_mode_2’, ‘alignments2D’]
[CPU: 2.09 GB] Loaded passthrough dset with 27429 items
[CPU: 2.09 GB] Intersection of output and passthrough has 27428 items
[CPU: 2.09 GB] Checking outputs for output group particles
[CPU: 2.09 GB] Updating job size…
[CPU: 2.09 GB] Exporting job and creating csg files…
[CPU: 2.09 GB] ***************************************************************
[CPU: 2.09 GB] Job complete. Total time 822.22s
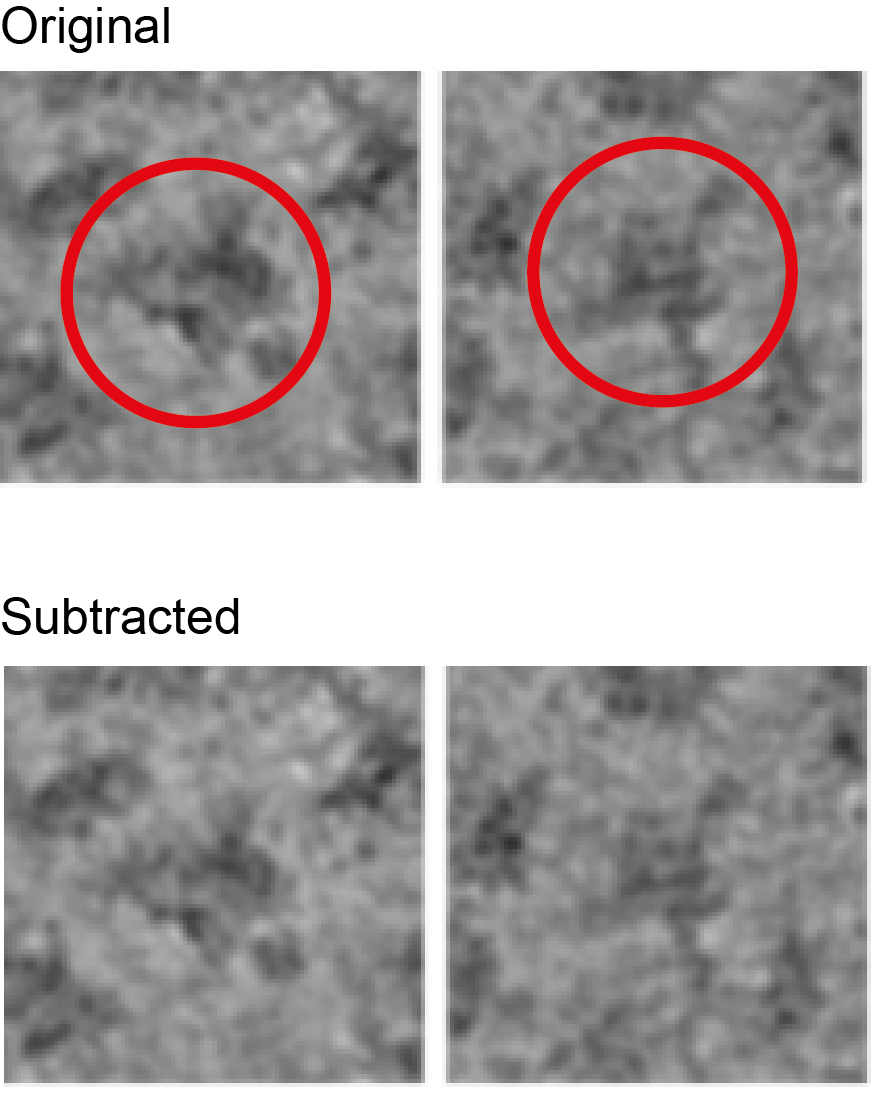
I am also attaching the particles. The 40S is circled in red, and should be substantially reduced (the mask covers about 80%).
Luca
Edit: now that I am blowing up the images, It looks as if the subtracted particles are overall lighter, but it seems not limited to the mask area.
Dear @cryo-lg,
Thanks for confirming the box sizes. It looks like something more subtle may be the cause of this. Could you first confirm that the mask is valued in [0,1] and not some other range? (Just in case – sometimes we’ve seen issues where imported masks have the wrong range).
If the mask is fine, my other guess would be that the particle scales are somehow mis-estimated and that is causing the poor scaling in subtraction. When you ran the previous refinement, did you turn on “Minimize over per-particle scale”? If you’re not sure, you can check the scale distribution in the input particle dataset by running a “Homogeneous Reconstruction Only” job – at the end, it will display a histogram of the particle scales. If scales aren’t all 1, it may be worth it to try re-refining the structure with the “Reset input per-particle scale” parameter on, and then trying subtraction again with the output particle stack.
The other potential cause I can think of is if the window inner/outer radii were changed during any of the processing. This can cause discrepancies with the multiplicative scale, so the subtraction job has to know what window radii were used in the previous refinement to correct the scale (some more info on the subtraction job is here).
Best,
Michael
1 Like
Hi Michael,
thank you for the reply.
- mask range: the mask was indeed ranged from -0.0325 to 0.4.
I noticed for the resizing in cryosparc I had left out the threshold option.
I re-ran my volume tools job, with threshold set to 0.05 this time. I downloaded the map, and chimera now showed a range of 0-1.
I re-ran the particle subtraction with this new mask and the particles looked similar to before.
So I ran a 2D classification, and yay!
I have a dark blotch in the center of the classes (see attached).
That is what I should get, correct?
As I checked everything:
2) Particle scales look ok.
3) Inner/outer radii are always the same (0.85 and 0.99 for the local refinement job with the subtraction mask, and for the subtraction job).
Looks like my mask was the problem.
Thanks a lot!
Kind regards,
Luca

1 Like
Dear @cryo-lg,
Glad to help! Looks like the mask was definitely the issue here. You can also check the subtraction by running a local refinement and checking for density in the subtracted region. Typically at this point, any residual density left over after subtraction would be due to suboptimal pose estimates, or heterogeneity/disorder (a recent discussion of this is here Particle subtraction of detergent belts still maintain low level signal of the belt).
Best,
Michael
2 Likes