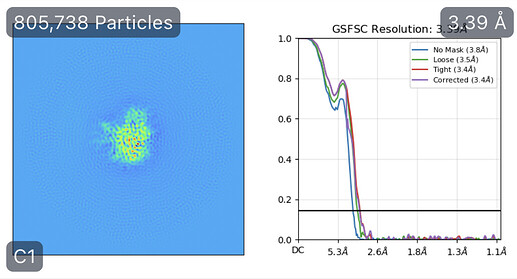
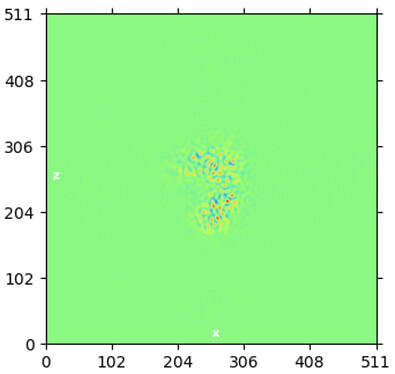
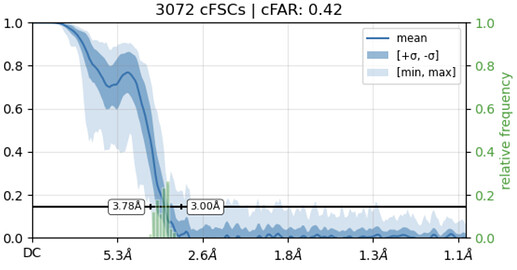
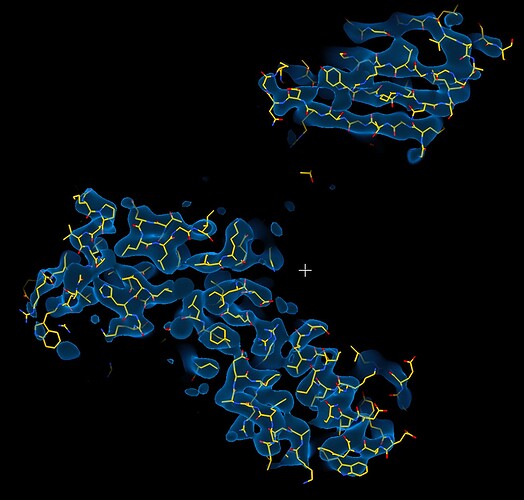
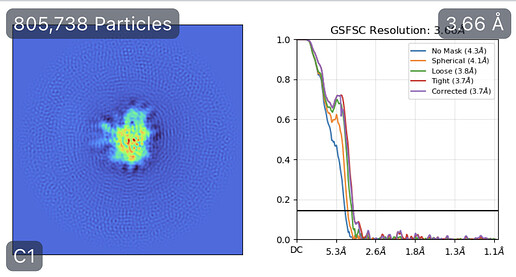
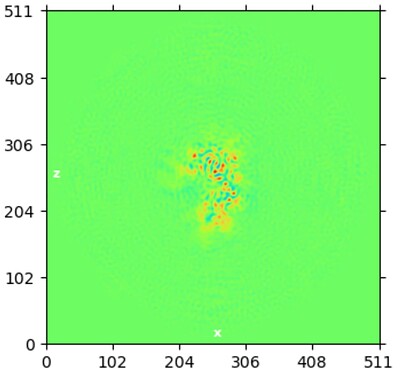
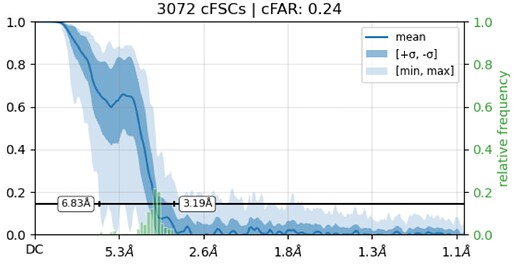
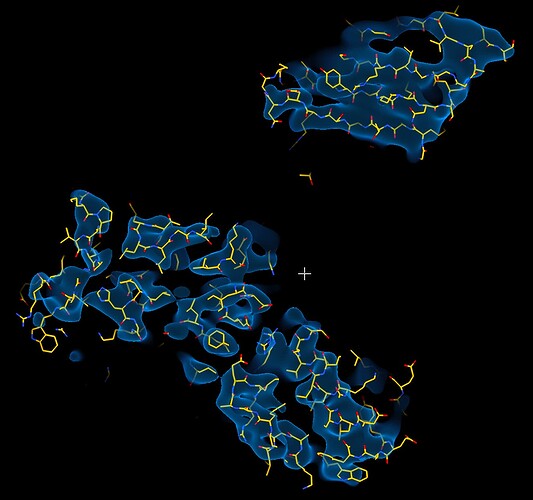
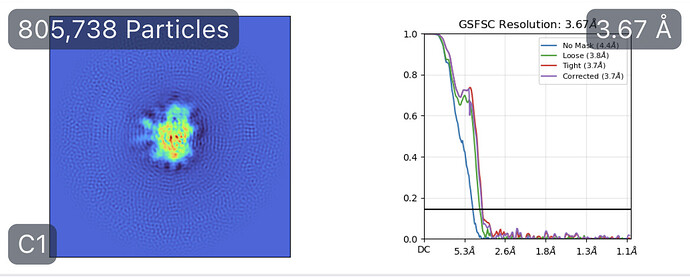
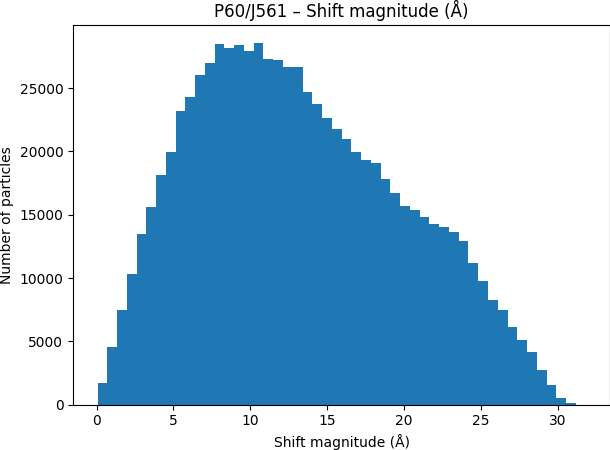
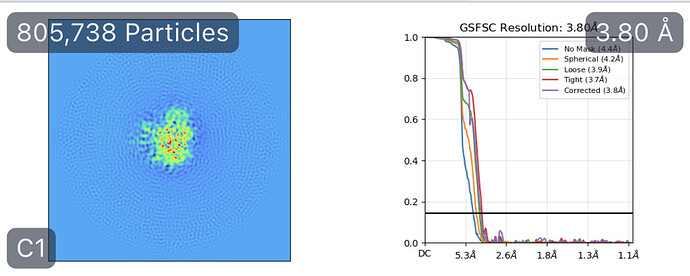
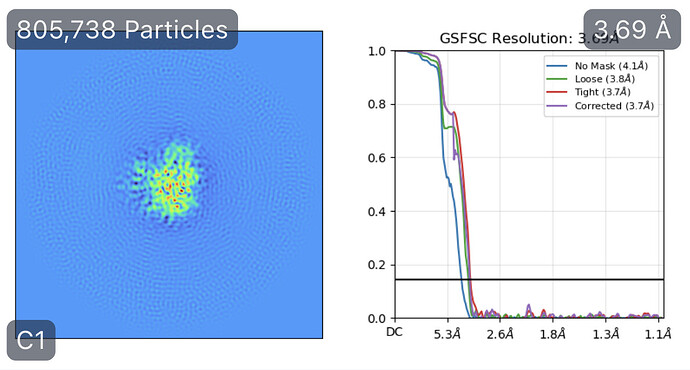
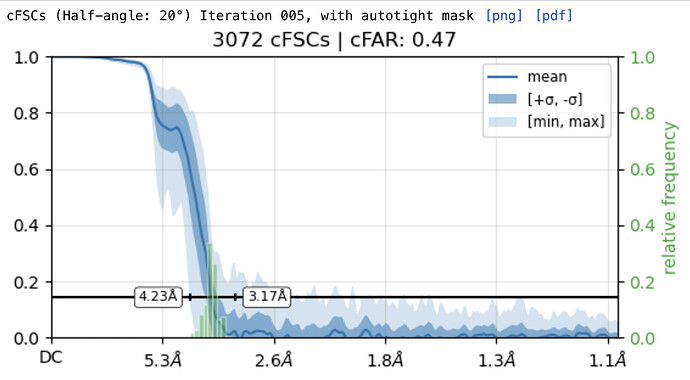
While investigating this I wrote (with assistance of ChatGPT) a CS tools script to plot the alignments3d/shift values (mostly to have an idea whether using them for recentering would be helpful).
Posting it here in case it is useful for anyone else (or to future-me!).
Script:
#!/usr/bin/env python3
"""
plot_shifts_external.py – raw-vs-converted shift histograms (constant psize)
Positional UIDs (any order, all required)
Pxx project
Wxx workspace
Jxx refinement job
Optional flag
--bins INT Histogram bin count (default 50)
"""
import argparse, json, sys
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
from cryosparc.tools import CryoSPARC
# ───────── helpers ─────────
def connect():
cfg = json.load(Path("~/instance_info.json").expanduser().open())
cs = CryoSPARC(**cfg)
assert cs.test_connection()
return cs
def classify(tokens):
out = {"P": None, "W": None, "J": None}
for tok in tokens:
tag = tok[0].upper()
if tag not in out:
raise RuntimeError(f"Bad UID '{tok}' (must start P/W/J)")
if out[tag]:
raise RuntimeError(f"Duplicate {tag} UID ('{out[tag]}' & '{tok}')")
out[tag] = tok
missing = [k for k, v in out.items() if v is None]
if missing:
raise RuntimeError("Missing UID(s): " + ", ".join(missing))
return out["P"], out["W"], out["J"]
def pixel_size(parts, job):
cols = [
"alignments3D/psize_A",
"microscope_parameters/psize_A",
"movies/psize_A",
"image_original_pixelsize_A",
"psize_A",
]
for col in cols:
if col in parts.fields():
return float(parts[col][0]), f"particle column '{col}'"
return float(job.load_output("volume")["map/psize_A"][0]), "refinement volume 'map/psize_A'"
def shift_px(parts):
if "alignments3D/shift_px" in parts.fields():
vec = parts["alignments3D/shift_px"]
elif "alignments3D/shift" in parts.fields():
vec = parts["alignments3D/shift"] # stored in px
else:
raise RuntimeError("No shift vector found.")
return np.linalg.norm(vec, axis=1)
def plot_hist(data, unit, title, bins):
fig = plt.figure()
plt.hist(data, bins=bins)
plt.xlabel(f"Shift magnitude ({unit})")
plt.ylabel("Number of particles")
plt.title(title)
plt.tight_layout()
return fig
# ───────── CLI ─────────
cli = argparse.ArgumentParser(description=__doc__,
formatter_class=argparse.RawDescriptionHelpFormatter)
cli.add_argument("uids", nargs="+", help="Three UIDs (P… W… J…) in any order")
cli.add_argument("--bins", type=int, default=50, help="Histogram bins")
args = cli.parse_args()
PUID, WUID, JUID = classify(args.uids)
# ───────── CryoSPARC data ─────────
cs = connect()
proj = cs.find_project(PUID)
job = proj.find_job(JUID)
parts = job.load_output("particles")
shift_px_vec = shift_px(parts)
apix, psrc = pixel_size(parts, job) # constant by assumption
shift_A_vec = shift_px_vec * apix
# ───────── External job ─────────
ext = proj.create_external_job(workspace_uid=WUID,
title=f"Shift histograms for {JUID}")
ext.add_input(type="particle", name="input_particles")
ext.connect("input_particles", JUID, "particles")
with ext.run():
# concise pixel-size info
ext.log(f"Pixel size used: {apix:.4f} Å/px (source: {psrc})")
# raw pixels
ext.log_plot(
plot_hist(shift_px_vec, "px",
f"{PUID}/{JUID} – Shift magnitude (raw pixels)", args.bins),
text=(f"Raw pixel shifts; N={len(shift_px_vec):,}, "
f"mean={np.mean(shift_px_vec):.2f} px, "
f"σ={np.std(shift_px_vec):.2f} px"),
formats=["png", "pdf"]
)
# converted Å
ext.log_plot(
plot_hist(shift_A_vec, "Å",
f"{PUID}/{JUID} – Shift magnitude (Å)", args.bins),
text=(f"Converted shifts (Å); N={len(shift_A_vec):,}, "
f"mean={np.mean(shift_A_vec):.2f} Å, "
f"σ={np.std(shift_A_vec):.2f} Å"),
formats=["png", "pdf"]
)
ext.log("Generated by plot_shifts_external.py (raw + Å)")
print(f"External job created: {ext.uid}")
Note instance_info.json needs to be in home dir, format:
Output: