Hi,
We have been having a problem where the particle extraction step hangs intermittently (sometimes this completes without errors) after cryosparc reaches the final micrograph This is happening for both versions 2.15.1 and 2.16.1 (installed on Ubuntu 16.04 on the master node and Centos 7 on the worker nodes). We have recently configured our system to have 3 worker nodes connecting to a master node in a non-cluster setup. All other jobs work in this configuration, except for this error with particle extraction.
I have checked the extract directory in the job directory and I can see particle stacks present for the micrographs.
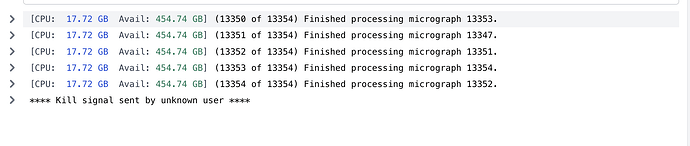
Here is the output for the extraction step.
[CPU: 3.56 GB] (5776 of 5784) Finished processing micrograph 5778.
[CPU: 3.56 GB] (5777 of 5784) Finished processing micrograph 5777.
[CPU: 3.56 GB] (5778 of 5784) Finished processing micrograph 5781.
[CPU: 3.56 GB] (5779 of 5784) Finished processing micrograph 5779.
[CPU: 3.56 GB] (5780 of 5784) Finished processing micrograph 5780.
[CPU: 3.56 GB] (5781 of 5784) Finished processing micrograph 5783.
[CPU: 3.56 GB] (5782 of 5784) Finished processing micrograph 5782.
Here is the cryosparc job log:
================= CRYOSPARCW ======= 2020-06-24 18:04:37.611009 =========
Project P36 Job J193
Master eagle Port 39002
===========================================================================
========= monitor process now starting main process
MAINPROCESS PID 30144
========= monitor process now waiting for main process
MAIN PID 30144
extract.run cryosparc2_compute.jobs.jobregister
========= sending heartbeat
========= sending heartbeat
***************************************************************
/app/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/skcuda/cublas.py:284: UserWarning: creating CUBLAS context to get version number
warnings.warn('creating CUBLAS context to get version number')
/app/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/skcuda/cublas.py:284: UserWarning: creating CUBLAS context to get version number
warnings.warn('creating CUBLAS context to get version number')
/app/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/skcuda/cublas.py:284: UserWarning: creating CUBLAS context to get version number
warnings.warn('creating CUBLAS context to get version number')
/app/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/skcuda/cublas.py:284: UserWarning: creating CUBLAS context to get version number
warnings.warn('creating CUBLAS context to get version number')
========= sending heartbeat
cryosparc2_compute/micrographs.py:405: RuntimeWarning: divide by zero encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
cryosparc2_compute/micrographs.py:405: RuntimeWarning: invalid value encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
cryosparc2_compute/micrographs.py:405: RuntimeWarning: divide by zero encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
cryosparc2_compute/micrographs.py:405: RuntimeWarning: invalid value encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
cryosparc2_compute/micrographs.py:405: RuntimeWarning: divide by zero encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
cryosparc2_compute/micrographs.py:405: RuntimeWarning: invalid value encountered in divide
return trim_mic(arr_zp_lp, out_shape) / trim_mic(ones_zp_lp, out_shape)
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
/app/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/matplotlib/pyplot.py:522: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).
max_open_warning, RuntimeWarning)
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
Let me know if you have any suggestions on ways to fix this problem.
Thanks