I have a dataset of a small protein that has a tendency to over-refine when adaptive marginalization is turned on. I have already removed duplicate particles from the dataset. Once the whole dataset is seen, and FSC-noise substitution is used, the corrected FSC resolution estimate agrees with the quality of the map. However, this causes some level of over-refinement during the first few steps of refinement and makes the refinement stop prematurely because the resolution decreases once FSC-noise substitution is used.
I can extend the refinement by increasing the number of final steps but I was wondering if it is possible to have FSC-noise substitution on from the first (or perhaps second) iteration? Alternatively, I assume using the full dataset from the beginning would also help with this problem. I tried adjust the “Initial batchsize” and “Batchsize epsilon” but could not get the desired behavior. I’ve included example FSC curves before and after the corrected FSC is calculated. Depending on the specific particle set used, the resolution estimate can go all the way to nyquist before FSC-noise substitution is used.
Any suggestions would be greatly appreciated!
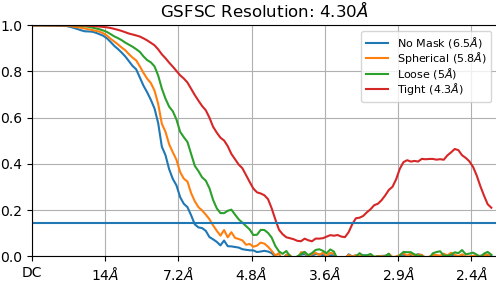
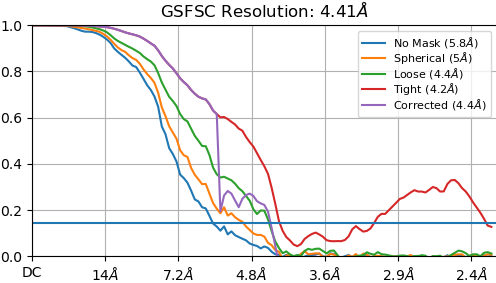
Thanks!
Evan