Dear All,
For " Case Study: Yeast U4/U6.U5 tri-snRNP", it needs files from EMPIAR-10072. In the directory for downloading in the webpage of EMPIAR-10072 (https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10073/), there were 2 kinds of files, one was “*.mrcs”, like "11Jul15_6124_cor_bin2_extractedparticles_11Jul15unchecked_shiny200-22Jun11Jul-7frames.mrcs ", one was the star file which can be extracted from “shiny_correctpaths_cleanedcorruptstacks.star.gz”.
From “https://guide.cryosparc.com/processing-data/tutorials-and-case-studies/case-study-yeast-u4-u6.u5-tri-snrnp” I am not clear whether the star file extracted from “shiny_correctpaths_cleanedcorruptstacks.star.gz”, or the series of *.mrcs files will be used for the Case study, or both, was the only file used by the Case study. I am expecting an explanation from you.
No matter which case, will you please let me know how to import the above required file for the Case study by cryoSPARC, which I mean by “Import Movies”, or by “Import Micrographs”, or by “Import Particle Stack”? It seems none works.
Or will you please tell me, how to have a completed job from cryoSPARC, from which I can drag the particles to job “Ab-Initio Reconstruction” as mentioned in the Case study?
I am looking forward to getting your reply.
Flemming
Hi @Flemming,
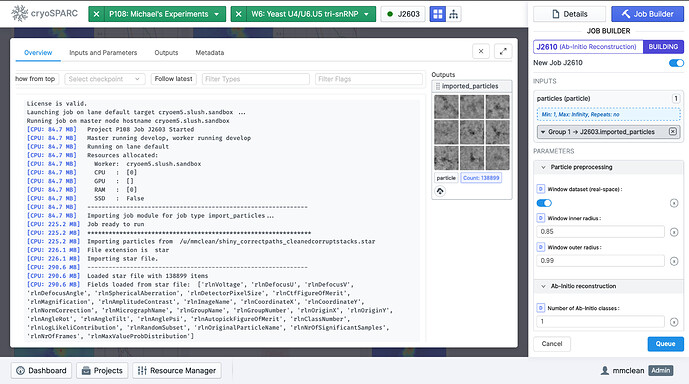
In this case study, we imported particles directly from EMPIAR entry 10073. In order to import these particles into cryoSPARC, you can do the following:
- Click on the “Job Builder” button on the top right of the desired workspace and build an “Import Particle Stack” job.
- In the “Particle meta path” parameter, fill in the full (i.e. absolute) path of the
shiny_correctpaths_cleanedcorruptstacks.star file.
- In the “Particle data path” parameter, fill in the full path of the directory that contains the
.mrcs files. If you downloaded directly from EMPIAR, this would probably be something like .../10073/data/
You can leave all other parameters of the Import Particles job as default, and then queue the job. The job should output a particle stack with 138899 particles in it.
From this, you can then build an Ab-Initio Reconstruction job and connect the particles outputted from the Import Particle Stack job into the Ab-Initio Reconstruction. When the ab-initio job is built, the particles can be connected by clicking on the Import Particle Stack job card, pressing the space bar, and then dragging over the imported_particles dataset into the particles input in the ab-initio job.
You can then launch the ab-initio job, followed by a homogeneous refinement, and continue on with the case study. Let me know if you have any other questions!
Best,
Michael
Dear All,
I have tried to input the EMPIAR entry 10073 related .star file and .mrcs files based on the advice of Michael. It really shows the particle stack contains 138899 particles. However the job “Import Partcle Stack” gives the following error message:
File “cryosparc_master/cryosparc_compute/run.py”, line 84, in cryosparc_compute.run.main
File “/home/user/mycryosparcv3/cryosparc_master/cryosparc_compute/jobs/imports/run.py”, line 352, in run_import_particles
hdr = mrc.read_mrc_header(mrc_file)
File “/home/user/mycryosparcv3/cryosparc_master/cryosparc_compute/blobio/mrc.py”, line 55, in read_mrc_header
[hdr[‘nx’], hdr[‘ny’], hdr[‘nz’], hdr[‘datatype’]] = header_int32[:4]
ValueError: not enough values to unpack (expected 4, got 0)
I am looking forward to getting your advice on how to have the job “Import Partcle Stack” successfully done for “Case Study: Yeast U4/U6.U5 tri-snRNP”.
Sincerely yours,
Flemming
Dear All,
The issue solved. The error message was caused by the mrcs files not properly downloaded. When the mrcs files were downloaded in linux, empiar-10073 datasets can be correctly recognized by cryosparc.
Flemming
1 Like