Hi, please forgive me if my question reflects a poor understanding of particle analysis- this is my first ever cryoEM dataset and analysis.
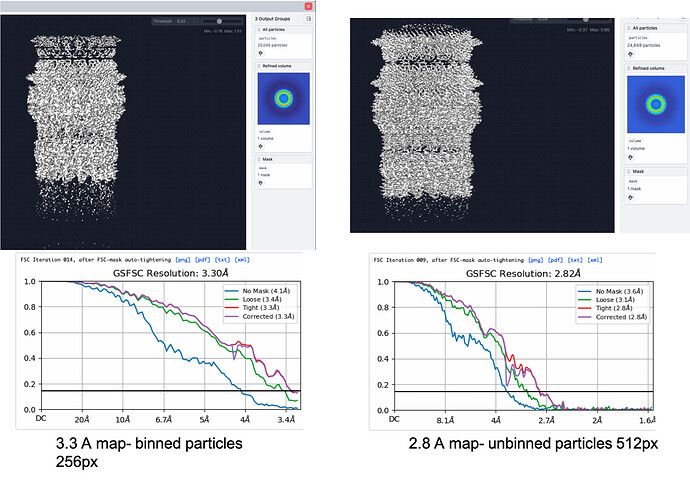
I used a subset of particles (25 k), extracted using a box size of 512, binned to 256 for ab-initio reconstruction, followed by one round of homogenous refinement, then used the output to run another homogenous refinement job applying the built in CTF refinement. I got a map at 3.3 angstroms.
I wanted to do the reconstruction with unbinned particles at 512 pixel size, so I re-extracted the 25 k particles without binning, and immediately used this as an input for a non-uniform refinement job (I wanted to see if I will get an improvement in my map if I used this type of refinement instead). I used the extracted particles as an input, and for the input map, I used the 3.3 angstroms one I got from the refinement of the binned particles.
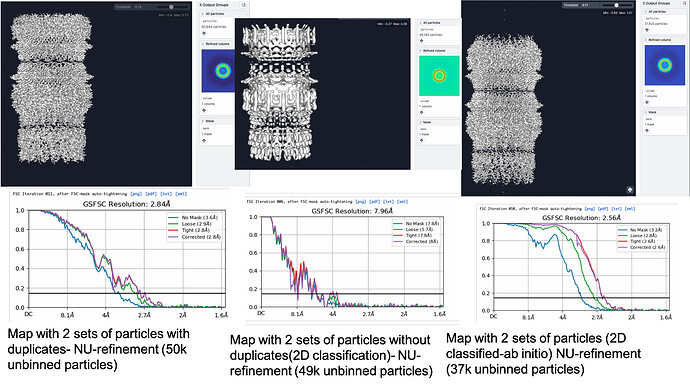
At the same time I could see I was lacking any good top views in my 25 k particles, so I re-extracted from the same micrographs (after applying some parameters to discard more of the bad ones and improve particle picks) a set of about 24 k particles that contained 500 of intact top views. I extracted them at the same box size of 512, and I want to add that in this case and in the unbinned re-extraction I mentioned above, I used an input from 2D classification so I believe the particles have 2D alignment information. Now for some reason, I added this 24 k particle set also to the input of the non-uniform alignment, not realizing at the time that there could be duplicate particles. The Job took a very long time, around 4 hours, running on two GPUs, and yielded a map of 2.8 angstroms.
When I had two problems : 1) there must be duplicates between my two particle subsets, 2) the input map had 3D alignment information from only half of the particles, I was extremely confused about how the map yielded that high resolution when I imagine it shouldn’t have worked. When I combined the particles stacks and used as an input for 2D classification, applying the “remove duplicates” parameter, I got only 1260 duplicates, so its a very large subset. However when I used this combined stack to repeat the non-uniform refinement (still using the same 3.3 angstrom map from the initial reconstruction) it yielded a very poor result, of about 8 angstroms. I believe this happened because combining the particles in new 2D classes messed up their alignment with the map I already had?..To avoid the mess, I tried running non-uniform refinement for each particle stack separately (but still with the same initial map input) and it seems to be doing better, but the run time is extremely long…given the stacks are just 25 k each…
To summarize this very long message I want to ask:
1- How could I get such a high resolution map when half the particles contributed non of the 3D alignment info in the input map?
2- How can I best combine these two particle stacks for a refinement job?
3- Why is non-uniform refinement taking a huge amount of time ~ 7 hours for such a low number of particles?
Thanks