I got the following error when trying to do exactly this: a custom segmentation .mrc file for a map with three segments.
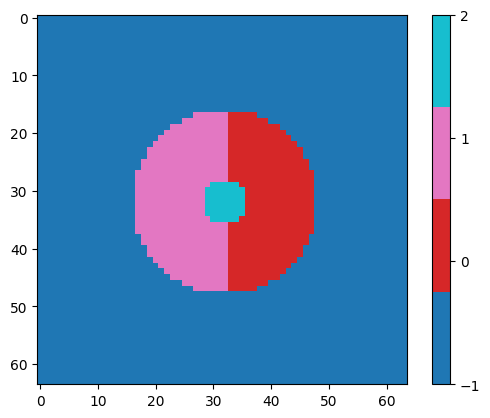
I created the segment mrc file with a custom script, and set the mrcfile datatype to np.int8
I set the solvent to -1.
When I set the segment voxels to to 0, 1, and 2, I kept seeing in the log that 3D Flex Mesh Prep only picked up on two segments [0,1].
When I set the segment voxels to 1, 2, and 3 I didn’t get this error anymore.
Loaded 3 segmentation with segment IDs: [0 1 2]
(But, using 2>1,2>3 to connect the segments didn’t work, so I did have to index the segments with 1>0,1>2 even though the values in the voxels weren’t the same.)
This appeared to be working alright (the segments look right in the viewer), but I got the following error.
Tree of segment connections:
> 1 > 0
> 2
Segment connections (segment indices): [(1, 0), (1, 2)]
Segmenting mesh...
Expanding input segments to fill mask...
Splitting contiguous mesh...
Fusing connected segments...
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 95, in cryosparc_master.cryosparc_compute.run.main
File "cryosparc_master/cryosparc_compute/jobs/flex_refine/run_meshprep.py", line 167, in cryosparc_master.cryosparc_compute.jobs.flex_refine.run_meshprep.run
File "cryosparc_master/cryosparc_compute/jobs/flex_refine/tetraseg.py", line 50, in cryosparc_master.cryosparc_compute.jobs.flex_refine.tetraseg.segment_and_fuse_tetra
File "cryosparc_master/cryosparc_compute/jobs/flex_refine/tetraseg.py", line 44, in cryosparc_master.cryosparc_compute.jobs.flex_refine.tetraseg.segment_and_fuse_tetra.points_from_cells
IndexError: arrays used as indices must be of integer (or boolean) type
Do I need to do something else to the .mrc, (e.g. the header?) file to ensure that the segments can be read in properly?