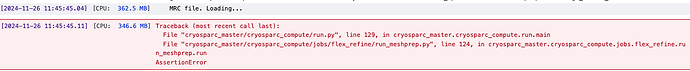
Hello, a following questioning on this:
I tried to make mesh by using individual protein pdb models by molmap then resample on the box size to my map of interested in chimera, then used the script below to change value of individual molmap volume so that any voxel > 0 is assigned to protein number (1,2,3, etc.) and -1 for voxel <0, I tried format either float32 or int8.
import mrcfile
import numpy as np
assign_val = 1
with mrcfile.open(volume_path, mode=‘r’) as mrc:
data = mrc.data.copy() # Create a copy to avoid modifying the original file
voxel_size = mrc.voxel_size
modified_data = np.where(data > 0, assign_val, -1).astype(np.int8)
with mrcfile.new(volume_path.replace(‘.mrc’, ‘_’ + str(assign_val) + ‘.mrc’), overwrite=True) as new_mrc:
new_mrc.set_data(modified_data)
new_mrc.voxel_size = voxel_size
volume_path = ‘~/J1061/J1061_n1.mrc’
Then, I merged individual maps with reassigned value using vop maximum command in chimera and resulted the volume with regions of interest at values 1,2,3,4… and solvent at -1, used this as the input of seg file. However, the mesh create job failed with Assertion Exception without further error message. If possible, some advice will be very helpful.