Hi,cryosparc team,
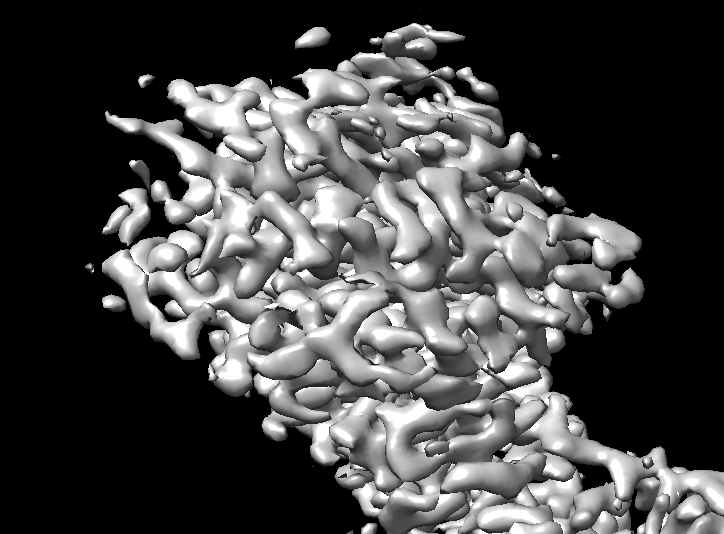
I have a doubt about my map’s extracellular feature.When I use crysoparc refine(no-uniform refinement had the same problem) my monomer data and dimer data,there is difference between monomer and dimer’s extracelluar domain.It’s not a structure difference ,it’s a map density problem.The dimer’s extracelluar domain’s density became abnomal even the resolution is 3A. You can look the screenshot.The dimer’s density become a slice and a slice,it is abnormal.Is there any interpretation,thanks very much.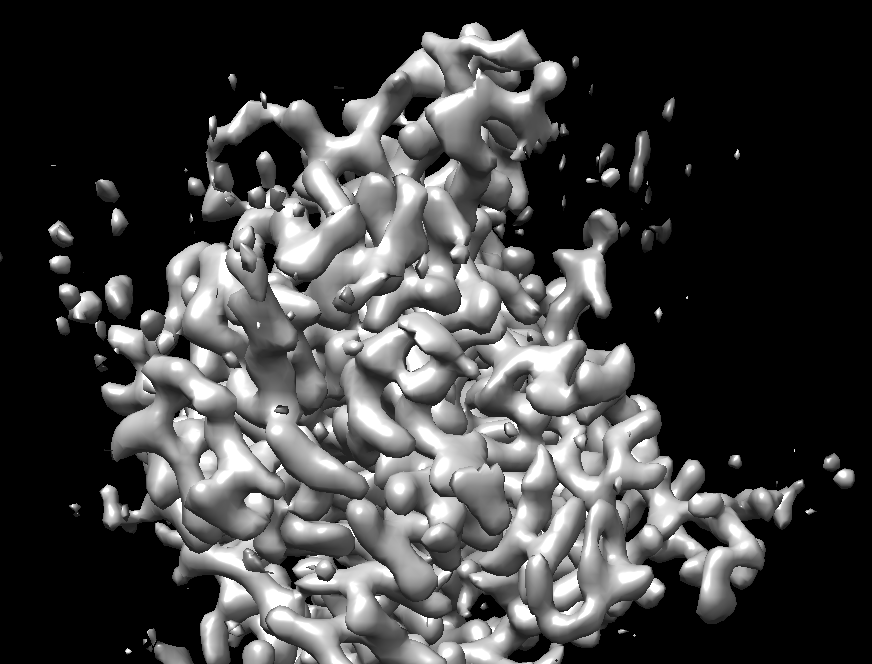
Hi @wonderful,
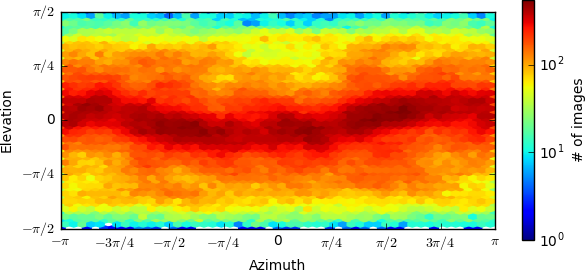
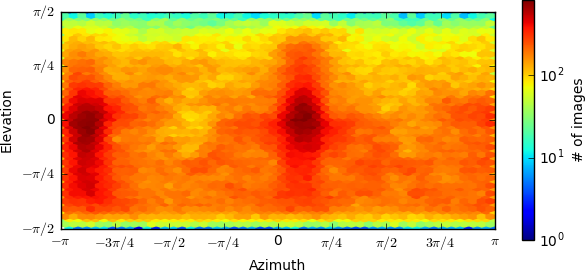
I’m guessing the first image is the monomer and the second the dimer? Is the abnormality you are referring to the fact that the dimer appears to be “stretched”? This looks like a common symptom of preferred orientation.
To confirm, the monomer and dimer datasets were collected separately, right? i.e. separate grids?
Can you post the orientation distributions from the two refinements?
Hi @apunjani,
Thanks very much for your reply.You are right,the first image is monomer and the second the dimer.it is seems “stretched”.I think maybe preferred orientation is the problem,is there any way to solve the problem.Can I use "rebalance 2D classes " job to solve this problem.I don’t very clear about this job’s usage.
The dimer and the monomer datasets were on one data set and one grid,them is two state in dataset.I classified them using heterorefinment.
Below is the orientation distributions from the two refinements(first one is monomer,second one is dimer),can you give me more suggestion?Thanks very much.
I also have another question.I don’t know the meaning “Posterior Precision Directional Distribution” plot,can you give some interpretation about the plot,thanks very much again.