Hi,
I’m not sure what has changed, but I consistently am not able to see an entire image without clipping in the shadows and highlights when I view micrographs in inspect particle picks or manual picker.
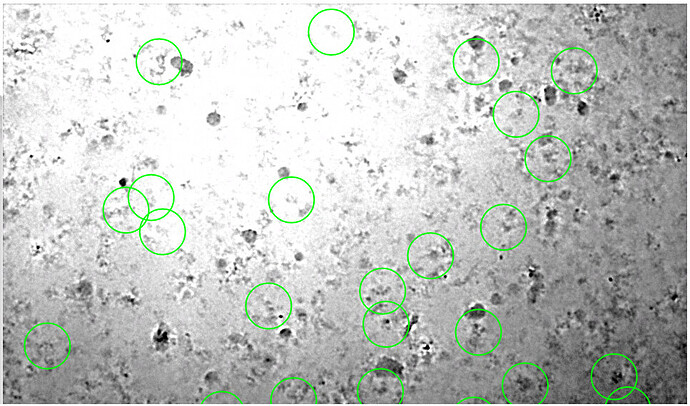
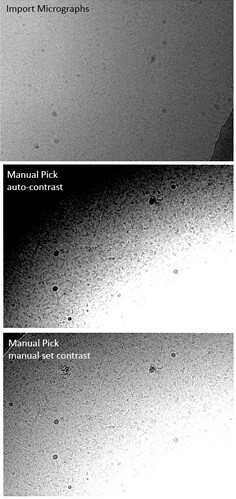
Using the override contrast allows me to tune the display settings for local regions of an image, but it is time consuming to repeat this process for every image. An example (denoised) micrograph is shown below, but the same is seen for the non-denoised images. Any ideas?
I also see this, with multiple datasets. It makes micrograph inspection or Manual Picking virtually impossible. In general, micrographs from tutorials like the 20S do seem to be visualised pretty good, but all our own datasets show this behaviour. Please see example below.
Even with manually adjusting the contrast, large areas of the micrographs remain inaccessible for manual picking.
We’ve noticed this with V3.0 and V3.1, but not with the V2.15, which was the last version we used before updating to V3.
I hope this can be fixed.
We see this as well but only (so far) with data collected in super resolution (on K3). Data collected in counting on K3 does not show this behaviour. @maarten.tuijtel @Ablakely does this tally with your experience?
My data were from a K2 camera in counting mode.
hmmm well scratch that theory I guess…
@sdawood would it be helpful to have examples of micrographs that reproduce this behavior as well as ones that don’t, in order to reproduce and fix?
We only see this in Manual Picker and Inspect Picks - Curate Exposures doesn’t show the same behavior
Hi @olibclarke @Ablakely @maarten.tuijtel
Could you let us know if the movies have been processed using either patch motion or rigid motion correction in cryoSPARC? If not, it could be the fact that micrographs are not background subtracted, which is causing the contrast normalization step to produce unhelpful previews. This contrast normalization step is only implemented in the manual picker and inspect picks job, which explains why exposure curation shows a different micrograph preview.
- Suhail
Ah that could well be the difference. The dataset that showed the behavior for me was processed with motioncor2 (relion implementation) as I want to do bayesian polishing later. Would it be possible to add a switch to turn contrast normalization off?
Cheers
Oli
Hey @olibclarke,
Yes, we could make contrast normalization an optional step. We will also implement automatic background subtraction for micrographs that have not been processed in patch/rigid motion correction.
- Suhail
My images were also motion corrected using Relion’s implementation, so that could very well be it.
-A
Hi, yes we generally use motioncor2 to correct for motion, and then load the outputs from that into cryosparc, so we generally don’t perform any motion correction in cryosparc itself.
Addition: we also see this behaviour on data from non-DED camera’s (yes, they still exist…), so they haven’t been motion-corrected at all.
Cheers, M
Hi Everyone,
This has been fixed in cryoSPARC v3.3.0 released December 1, 2021.
See: cryosparc.com/updates