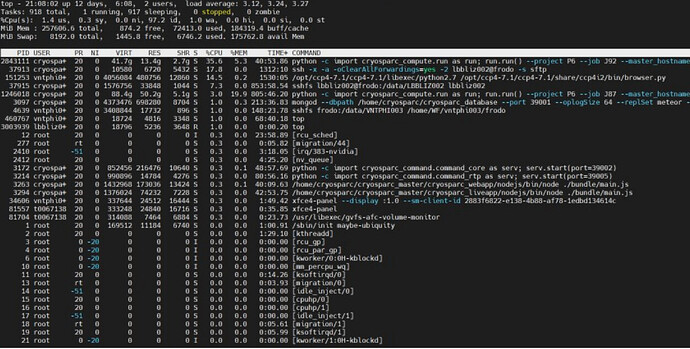
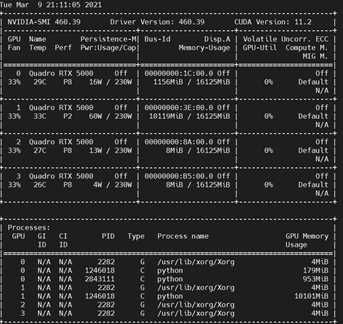
Hi, I am getting “memory error” at Iteration 17 when I run local refinement for 400px box with NU-refinement on and Fulcrum coordinates. The same job w/o Fulcrum coordinates was completed properly. Also, the similar jobs with different volume and mask with NU-refinement ON and Fulcrum coordinates were successfully completed. was completed. I’m using v2.14.2 (24 cores, 64GB RAM, 2GPUs with SSD). Could somebody help me solve this issue?
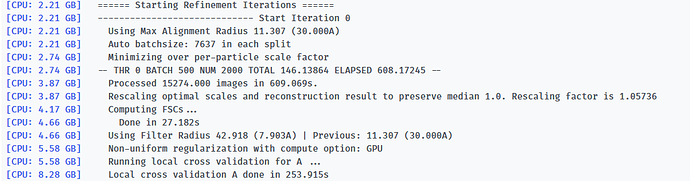
Here is the log of Iteration 17 and the error message.
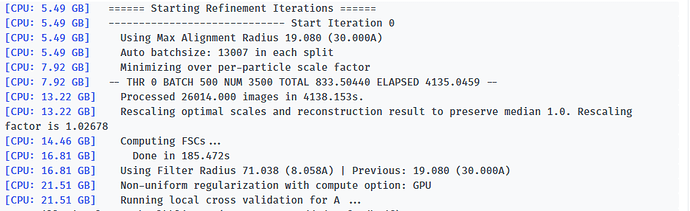
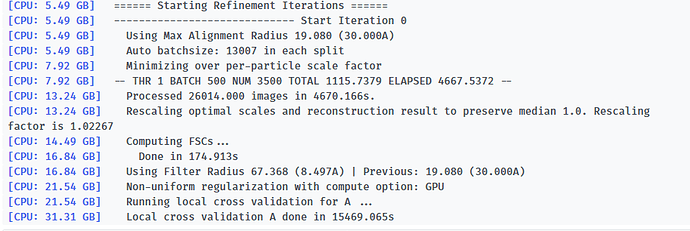
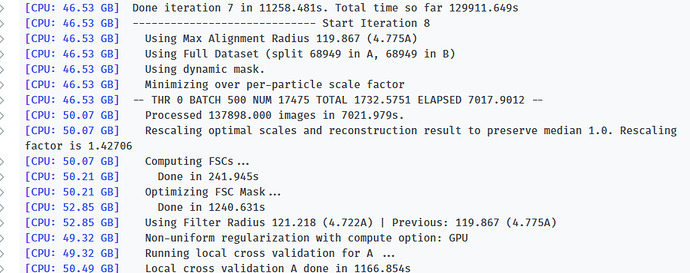
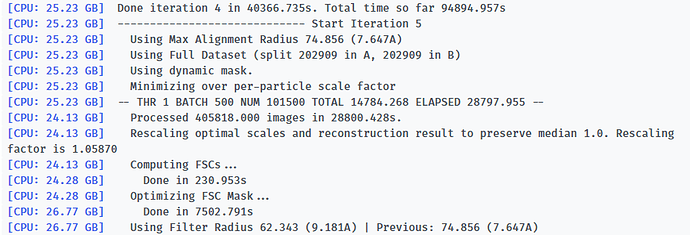
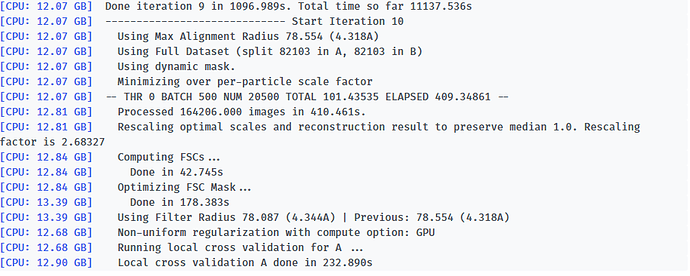
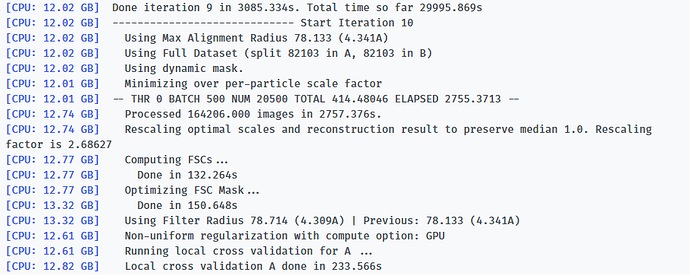
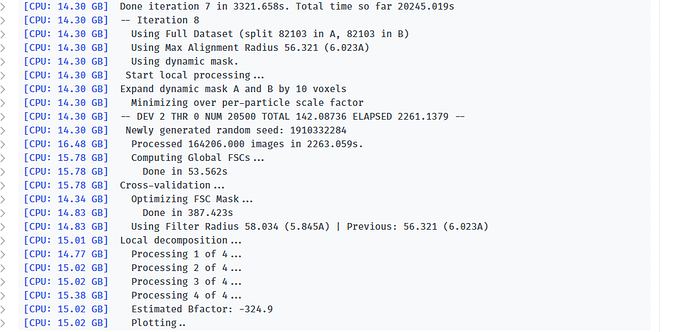
-- Iteration 17
[CPU: 48.33 GB] Using Full Dataset (split 50316 in A, 50316 in B)
[CPU: 48.33 GB] Using Max Alignment Radius 112.437 (3.664A)
[CPU: 49.29 GB] Using previous iteration scale factors for each particle during alignment
[CPU: 49.29 GB] Current alpha values ( 1.00 | 1.00 | 1.00 | 1.00 | 1.00 )
[CPU: 49.29 GB] Using best alpha for reconstruction in each iteration
[CPU: 49.29 GB] -- DEV 0 THR 1 NUM 12658 TOTAL 87.310928 ELAPSED 358.76506 --
[CPU: 52.14 GB] Processed 100632 images in 4562.842s.
[CPU: 52.62 GB] Computing FSCs...
[CPU: 52.62 GB] Done in 48.447s
[CPU: 52.62 GB] Optimizing FSC Mask...
[CPU: 38.99 GB] Traceback (most recent call last):
File "cryosparc2_worker/cryosparc2_compute/run.py", line 82, in cryosparc2_compute.run.main
File "cryosparc2_worker/cryosparc2_compute/jobs/local_refine/run.py", line 586, in cryosparc2_compute.jobs.local_refine.run.run_naive_local_refine
File "cryosparc2_compute/sigproc.py", line 1025, in find_best_fsc
tight_near_ang=near, tight_far_ang=far, initmask=initmask)
File "cryosparc2_compute/sigproc.py", line 1001, in compute_all_fscs
radwns, fsc_true, fsc_noisesub = noise_sub_fsc (rMA, rMB, mask, radwn_noisesub_start, radwn_max)
File "cryosparc2_compute/sigproc.py", line 823, in noise_sub_fsc
fMBrand = fourier.fft(fourier.ifft(randomphases(fourier.fft(MB), radwn_ns)) * mask)
File "cryosparc2_compute/fourier.py", line 110, in fft
return fftcenter3(x, fft_threads)
File "cryosparc2_compute/fourier.py", line 74, in fftcenter3
fv = fftmod.fftn(tmp, threads=th)
File "/home/owner/cryosparc2c/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/pyfftw/interfaces/numpy_fft.py", line 183, in fftn
calling_func, normalise_idft=normalise_idft, ortho=ortho)
File "/home/owner/cryosparc2c/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/pyfftw/interfaces/_utils.py", line 125, in _Xfftn
FFTW_object = getattr(builders, calling_func)(*planner_args)
File "/home/owner/cryosparc2c/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/pyfftw/builders/builders.py", line 364, in fftn
avoid_copy, inverse, real)
File "/home/owner/cryosparc2c/cryosparc2_worker/deps/anaconda/lib/python2.7/site-packages/pyfftw/builders/_utils.py", line 186, in _Xfftn
input_array = pyfftw.byte_align(input_array)
File "pyfftw/utils.pxi", line 96, in pyfftw.pyfftw.byte_align
File "pyfftw/utils.pxi", line 130, in pyfftw.pyfftw.byte_align
File "pyfftw/utils.pxi", line 172, in pyfftw.pyfftw.empty_aligned
File "pyfftw/utils.pxi", line 201, in pyfftw.pyfftw.empty_aligned
MemoryError