Hi Kye, after trying this myself I have a couple more queries/thoughts:
a. In step 5, is it neccessary to actually re-extract? Won’t the updated coordinates be sufficient for apply traj? I am trying it at the moment and it seems to be working without extracting after VAT, but apply traj doesn’t give any intermediate outputs, or plot the subparticle trajectories or particle centers on the micrographs, so it is hard to be sure.
Also, I’m not sure if using re-extracted particles is even compatible with Apply Traj if Fourier cropping was initially performed during Patch Motion (as Apply Traj seems to fail if the box size differs between the Apply Traj value (which is based on the original movie pixel size) and the input particles)
b. It would be very useful to have an option to Fourier Crop in apply trajectories. Currently this workflow leads to giant, disk-filling particle stacks if one is using super-res movies with already large particles and symmetry expansion, making this workflow unfortunately impractical for some of the cases we would like to try it on. It would also be helpful if apply trajectories had a progress bar like RBMC - it is currently difficult to estimate how long it will take to complete.
c. Apply Traj produces a bunch of unlabeled plots at the start of the log. What do these signify?
Also, Apply Traj has an option “produce motion diagnostic plots for n movies”, but it does not produce motion diagnostic plots for any movies, regardless of what value this is set to.
d. Apply Traj seems to be running and processing movies, but some movies (just a handful so far) seem to be failing with CUDA allocation errors. This is on a 3090, initial box size 1200 (super-res). Is this something to be concerned about? Can the incomplete movies be reprocessed later? I guess this also indicates that larger box sizes will be impractical with any card?
Error occurred while processing J54/imported/007171423974844969476_24nov04c_grid1_100_00006gr_00077sq_v03_00005hln_00027enn.frames.tif Traceback (most recent call last): File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 851, in _attempt_allocation return allocator() File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 1054, in allocator return driver.cuMemAlloc(size) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 348, in safe_cuda_api_call return self._check_cuda_python_error(fname, libfn(*args)) File
“/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 408, in _check_cuda_python_error raise CudaAPIError(retcode, msg) numba.cuda.cudadrv.driver.CudaAPIError: [CUresult.CUDA_ERROR_OUT_OF_MEMORY] Call to cuMemAlloc results in CUDA_ERROR_OUT_OF_MEMORY During handling of the above exception, another exception occurred: Traceback (most recent call last): File “/home/exx/cryosparc/cryosparc_worker/cryosparc_compute/jobs/pipeline.py”, line 59, in exec return self.process(item) File “cryosparc_master/cryosparc_compute/jobs/motioncorrection/run_applytraj.py”, line 253, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.run_applytraj.run_apply_traj_multi.motionworker.process File “cryosparc_master/cryosparc_compute/jobs/motioncorrection/run_applytraj.py”, line 261, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.run_applytraj.run_apply_traj_multi.motionworker.process File “cryosparc_master/cryosparc_compute/jobs/motioncorrection/run_applytraj.py”, line 262, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.run_applytraj.run_apply_traj_multi.motionworker.process File “cryosparc_master/cryosparc_compute/jobs/motioncorrection/motioncorrection.py”, line 742, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.motioncorrection.motion_correction_apply_traj_patches File “cryosparc_master/cryosparc_compute/jobs/motioncorrection/motioncorrection.py”, line 763, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.motioncorrection.motion_correction_apply_traj_patches File “cryosparc_master/cryosparc_compute/gpu/gpucore.py”, line 398, in cryosparc_master.cryosparc_compute.gpu.gpucore.EngineBaseThread.ensure_allocated File “/home/exx/cryosparc/cryosparc_worker/cryosparc_compute/gpu/gpuarray.py”, line 376, in empty return device_array(shape, dtype, stream=stream) File “/home/exx/cryosparc/cryosparc_worker/cryosparc_compute/gpu/gpuarray.py”, line 332, in device_array arr = GPUArray(shape=shape, strides=strides, dtype=dtype, stream=stream) File “/home/exx/cryosparc/cryosparc_worker/cryosparc_compute/gpu/gpuarray.py”, line 127, in init super().init(shape, strides, dtype, stream, gpu_data) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/devicearray.py”, line 103, in init gpu_data = devices.get_context().memalloc(self.alloc_size) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 1372, in memalloc return self.memory_manager.memalloc(bytesize) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 1056, in memalloc ptr = self._attempt_allocation(allocator) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 863, in _attempt_allocation return allocator() File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 1054, in allocator return driver.cuMemAlloc(size) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 348, in safe_cuda_api_call return self._check_cuda_python_error(fname, libfn(*args)) File “/home/exx/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/numba/cuda/cudadrv/driver.py”, line 408, in _check_cuda_python_error raise CudaAPIError(retcode, msg) numba.cuda.cudadrv.driver.CudaAPIError: [CUresult.CUDA_ERROR_OUT_OF_MEMORY] Call to cuMemAlloc results in CUDA_ERROR_OUT_OF_MEMORY Marking as incomplete and continuing…
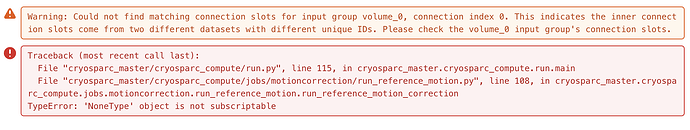
e. If one attempts to only process a subset of movies (in this case 50), Apply Traj fails with an error:
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 129, in cryosparc_master.cryosparc_compute.run.main
File "cryosparc_master/cryosparc_compute/jobs/motioncorrection/run_applytraj.py", line 340, in cryosparc_master.cryosparc_compute.jobs.motioncorrection.run_applytraj.run_apply_traj_multi
File "/home/exx/cryosparc/cryosparc_worker/cryosparc_tools/cryosparc/dataset.py", line 1487, in mask
assert len(mask) == len(self), f"Mask with size {len(mask)} does not match expected dataset size {len(self)}"
AssertionError: Mask with size 50 does not match expected dataset size 10438