Hi, I collected a dataset on a Krios/K2, 40 frames, 0.55A/pix (super-resolution). The micrographs were saved in 4-bit encryption. I used newstack to convert them into 8-bit encryption, imported them into cryosparc with a supposedly correct gain reference file, then did patch motion correct (default params) and patch ctf estimation (default parameters).
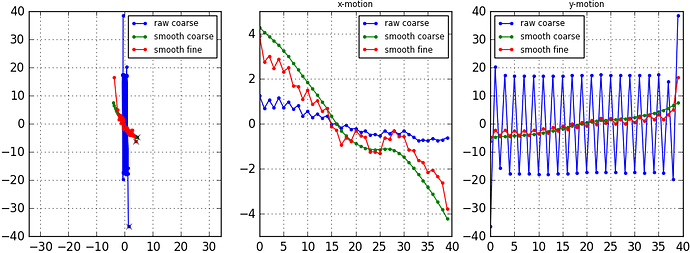
There seems to be DRAMATICALLY large motions.
In the manually curation jobs the “total drift” plot shows some drifts in the range of 1000 pixels! The pixel size I used is 0.55A (super-resolution). This would seem to indicate that there are drifts almost at micron level! Am I interpreting this correctly?
Estimated CTF resolution is <5A for the majority of the micrographs.
Any suggestions as to what is going on here? Was there data collection messed up here? Or may be something incorrect that I am doing with patch motion correction?
When I imported the micrographs, there seems to be some indication that gain correction file was off - I see patterns of dark lines at the same locations across several micrographs (may be energy filter alignment was off). Could that have led to this pattern of motions/high total full frame motion? If so, can anything be done about it?