Hi all,
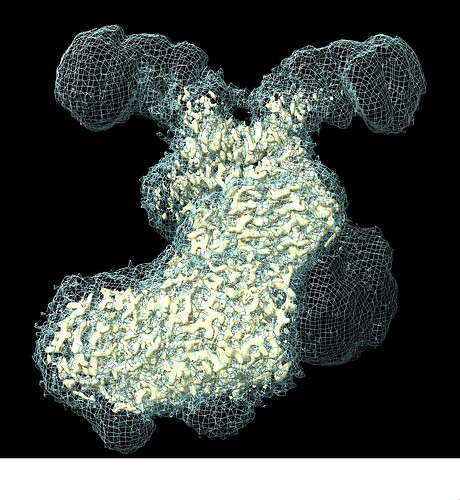
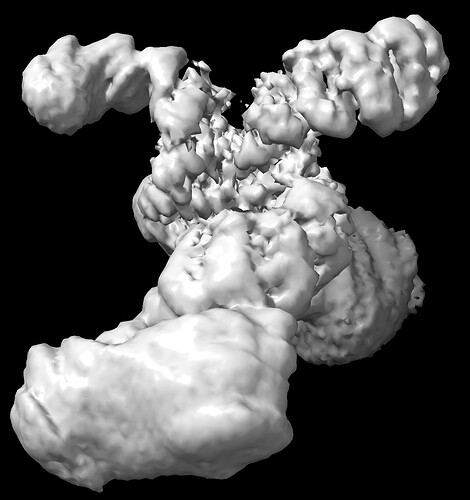
I have been trying to characterize the structure of a homodimeric protein (~110 kDa per monomer) with a superhelical domain bound to a peptide that extends through that superhelix. I have followed a standard pipeline of particle picking, heterogeneous refinement to weed out junk, and nonuniform refinement of ~2m particles to get an initial structure, which is around 2.4A resolution and doesn’t display too much orientation bias. However, examination of the resultant maps shows that the N-terminal end of the superhelix is poorly aligned and details can not be resolved. I have attached an image below of the sharpened map at a reasonable model-building contour and the unsharpened map contoured to show the ‘full’ protein. The region in question is at the top right, and the point of dimerization is at the base of the low-resolution regions.
I’ve tried various means of resolving this region of the protein, which I will summarize below:
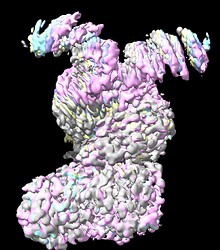
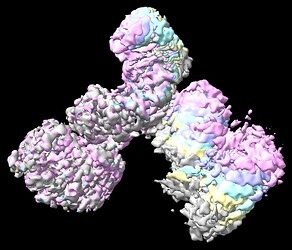
Local refinement using various masks around a single N-terminal domain helped somewhat, but was not a silver bullet. I’ve attached an image below showing the resultant unsharpened map from a representative job. Of note, local refinement did reveal presence of part of the interacting peptide bound in the central lumen of the N-terminus, which was not observed before. However, the N-terminal helices were still not at a quality where sidechain density can be modelled.
I also tried 3DVA and 3DFlex analysis, which indicated significant flexibility in the complex about the dimerization point in both the second monomer and both N-termini.
front view of 3Dflex results
side view of 3D flex results
I tried to follow a 3D classification scheme outlined in the recent end-to-end GPCR processing guide to classify my particles based on volumes from the 3D flex output, but so far this has not resulted in any notable density improvements in the N-termini, and in fact has in some ways worsened the density and created secondary problems in the previously well-aligned C-terminal domain.
I’m very interested in resolving the N-terminal density as I believe it may explain some in vitro observations I have made, but I’m running out of ideas for troubleshooting. Does anyone have any suggestions to resolve this density?