normalisation issue?
That’s what I’m thinking!
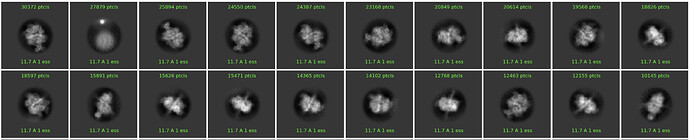
The 2D classes I posted are from the cryosparc extracted particles. Here is the 2D class from Warp extracted particles (this is from a larger particle stack, which includes the subset we’ve been discussing):
This does contain a similar class near top left. Is this a hot pixel/defect?
Here is the refinement again with dynamic mask near at 20A and dynamic mask far at 40A:
The results are worse compared to the tighter dynamic mask.
Thanks,
Matt
Please try to clean the dataset, only keep shiny classes and make a reconstruction at bin4.
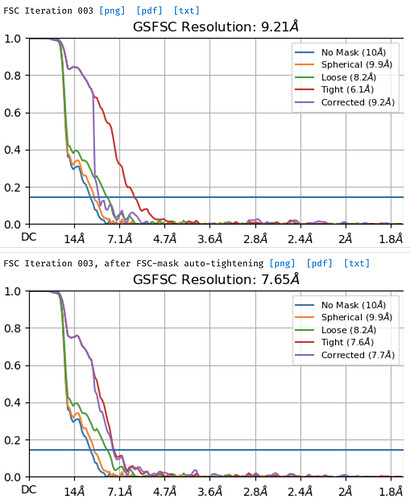
You should easily reach Nyquist and get a decent fsc curve.
If that works, go to bin2 and reconstruct again.
If your 2D classes from warp picking are unbinned, I’m wondering why the resolution is limited to ~ 11A.
Yeah agree - I definitely would not keep classes like the second most populated one you’ve posted here, yes looks like a hot pixel. More cleanup may help. Do you see white speckles on the micrographs if you look closely?
It would be nice to sit in a room with all of you to better hash this out:)
The stack extracted from Relion/Warp was cleaned originally which led to 4.73/4.91A map (see top of post). My goal was to take this stack to perform local motion correction in cryosparc. So I needed to re-link and re-extract particle coordinates with cryosparc processed movies. I was hoping the refinement of the same particles would yield the same results, but this wasn’t the case (5.77/7.11A and strange). Also, Refinement in Relion of this particle stack looks fine.
My strategy for cleaning particle stacks is:
- 5 class ab initio on clean 2d classes
- 5 class heterogenous refinement on all particles
- homo refinement on particles of interest (led to 4.73/4.91A map and these are particles coordinates I wanted to transfer for extraction within cryosparc)
My point here is (regardless of my dirty particle stack) extracting particles from micrographs processed in cryosparc yields refinement results significantly different from the same particles extracted from micrographs processed elsewhere. Maybe this is a normalization issue which is altering how cryosparc weights these particles?
Btw, I’ve now cleaned up the cryosparc extracted particles with a 3 class ab initio and the final refinement looks normal, but resolution is lost (5.04A/6.03A). Having to clean again kind of defeats the purpose of transferring the particles.
At least for now, maybe its best to start from scratch and re-process all particles from the micrographs processed in cryosparc.
Thanks,
Matt
your initial refinement (the “good”) refinement doesn’t really look normal though - there are clear masking artifacts and overfitting streaks. We often see this when the initial particle stack is still a bit dirty - and the 2D classes you posted from this stack seem to reflect this (the second most populated class has a big bright artefact).
For cleaning up, I often find heterogeneous refinement using “decoy” classes - random density - and one predefined good class works well to remove junk.
I agree it is weird though…
Cheers
Oli
I do agree that the particles are a bit messy and I’ll try your decoy strategy as I remember you discussing this in the past. Currently, I use the bad classes from ab initio as “decoys” in heterogenous refinement, but maybe your method yields better results.
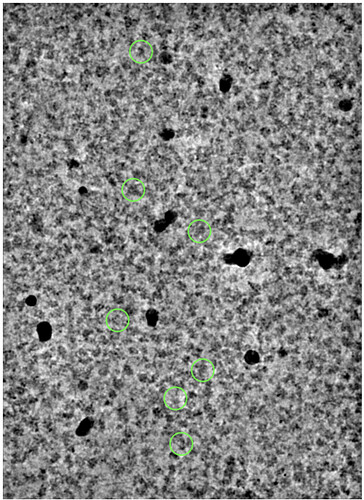
And I’m also considering removing those hot pixel classes in 2D classification. I wanted to inspect these particles with the Inspect particle picks job to see if they were all in the same location in different micrographs, but this isn’t possible b/c there are no pick_stats which you’ve posted on.
Do any of you use the defect files for processing? I notice these fields have showed up in all the processing softwares recently. If so, how do you generate the txt (cryosparc) or mrc (relion/warp) defect files?
Thanks,
Matt
re the pick stats - you can get around this IIRC by using the particles as input for a manual picking job, rather than inspect picks
Starting from scratch would definitely be faster than finding out what causes that behavior.
As stressed out already, despite the better nominal resolution the refinement from warped particles was suspicious as well.
I never used a defects file, would only make sense if that pixel error is constant in all your images.
First check this before hassling around with that…
cheers
The coordinates are very random for this class:
Seems to have something to do with sample then. Could be DNA since it is present.
And, yes, I’ll start from scratch with initial particle stack. I’m now wary of moving subsets of particle stacks between softwares.
Best,
Matt
These hot pixels we have seen in the past with the K3 (I call them hot pixels but actually I think they are just pixels where the gain ref is saturated). You can often remove them by applying the remove_outlier_pixels utility in CisTEM to the gain reference. This will often improve motion correction, too.