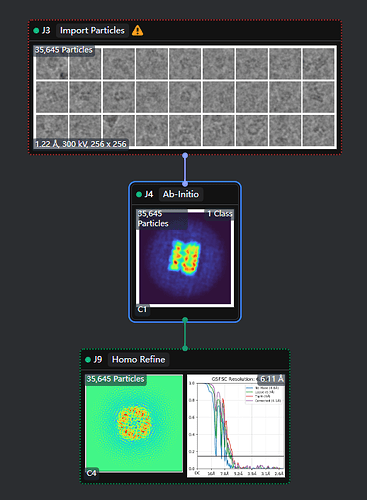
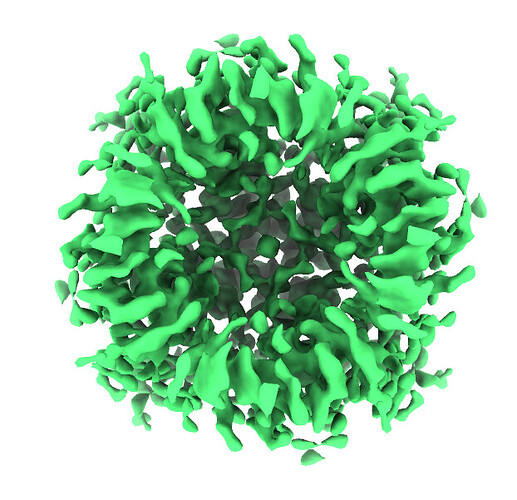
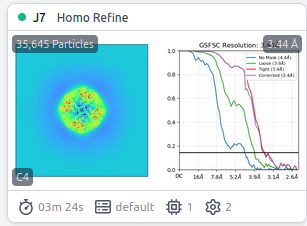
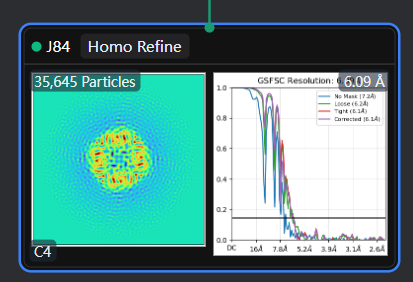
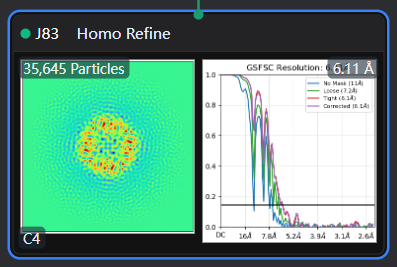
The reconstruction result of EMPIAR-10005 should be below 4 Å, but after performing a simple ab-initio reconstruction and a single homogeneous refinement with C4 symmetry, the result is around 6.1 Å. And here is my workflow.
Is there anything wrong with my data processing step ?
The sharp dips in the FSC make me think uncorrected beam tilt. If the original data was processed with RELION, the way CryoSPARC describes its CTF differs enough that you may need to re-calculate the global CTF.
Thanks for your reply ! This may be the reason of the problem.
I directly download the particle data from EMPIAR database, and the data was processed from RELION.
I’m not familiar with this software, do you have any idea how to re-calculate the global CTF, should I import an Movies data or Micrographs and then run the CTF Estimation? Or can I just calculate the global CTF from the particle stack?
Sorry to bother you again.
The sharp dips in the FSC make me think uncorrected beam tilt. If the original data was processed with RELION, the way CryoSPARC describes its CTF differs enough that you may need to re-calculate the global CTF.
I would think in this case poor convergence from a crappy ab initio might be more likely - this is the TRPV1 dataset from Yifan Cheng’s lab, originally published 2013 - no beam tilt corrections applied during original processing I think.
If I recall there is some beam tilt, and correcting it allows you to improve from 3.2 to 2.8ish, but it is not so severe as to curtail resolution at 6Å.
What parameters are you using for ab initio & refinement? Some tweaks may be required.
And how did the ab initio look in 3D - did it look like TRPV1, and isotropic? Or was it stretched or strange looking?
For refinement, I would change the initial lowpass to 15 - sometimes it can get stuck at low res… and for a better ab initio, try starting at 12 Å and finishing at 9 Å (although you are right that this looks ok, so I’m surprised it is giving you trouble).
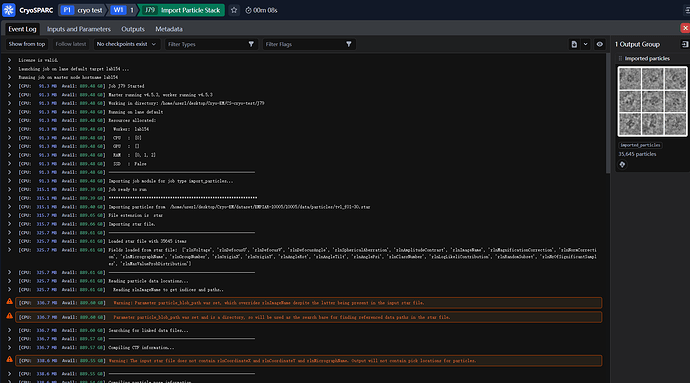
The exclamation mark indicates that there was a warning during import - what was it warning about? Just absent particle locations, or any errors re import of CTF params?
Possible. I’ve just looked at the EMPIAR dataset and the particle stacks come in lots of different flavours - all frame sum, 3-16 frame sum, individual frames - what stack is used will have a big effect.
Now I’m curious. Let’s have a play. ![]()
edit: The .star file on PDBJ EMPIAR doesn’t even reference the stack filename; it’s tv1.mrcs in the .star, but the stacks are different names. Either CryoSPARC did something automatically (possibly relating to that yellow exclamation mark on the Import) or there is the possibility that a non-optimal stack was used.
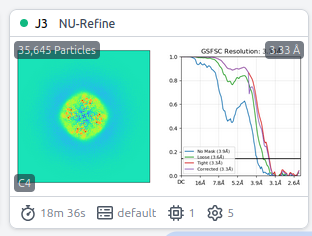
A dumb reconstruct gets 3.84Ang from the 1-30 frame stack… NU refine running now. Fresh ab initio looks good with the same stack as well.
@RIA1159; what particle stack did you use during import?
edit: From 1-30 stack:
NU refine:
Homogeneous refine:
edit 2:
It gets more interesting… first 100 mics (motion corrected stack) are K2 dimensions (3710x3738) but all the rest of them are square (3710x3710) which sends CryoSPARC’s extraction routines into spasms:
Error occurred while processing micrograph J5/imported/017598834880323366821_stack_0394_2x_SumCorr.mrc
Traceback (most recent call last):
File "/home/cryosparcer/bin/cryosparc_worker/cryosparc_compute/jobs/pipeline.py", line 59, in exec
return self.process(item)
File "/home/cryosparcer/bin/cryosparc_worker/cryosparc_compute/jobs/extract/run.py", line 510, in process
result = extraction_gpu.do_extract_particles_single_mic_gpu(mic=mic, bg_bin=bg_bin,
File "/home/cryosparcer/bin/cryosparc_worker/cryosparc_compute/jobs/extract/extraction_gpu.py", line 54, in do_extract_particles_single_mic_gpu
assert mic_shape == mic.shape, "Micrograph shapes are inconsistent!" # otherwise micrograph_blob/shape is inconsistent with the actual frame shape
AssertionError: Micrograph shapes are inconsistent!
Marking J5/imported/017598834880323366821_stack_0394_2x_SumCorr.mrc as incomplete and continuing...
Will raise as separate post.
It‘s so wreid, I tried both ‘tv1_f01-30.mrc’ with summing frame from 1 to 30 and ‘tv1_f03-16.mrc’ with frame from 3 to 16, but the reconstruct all gets near 6.0Ang.
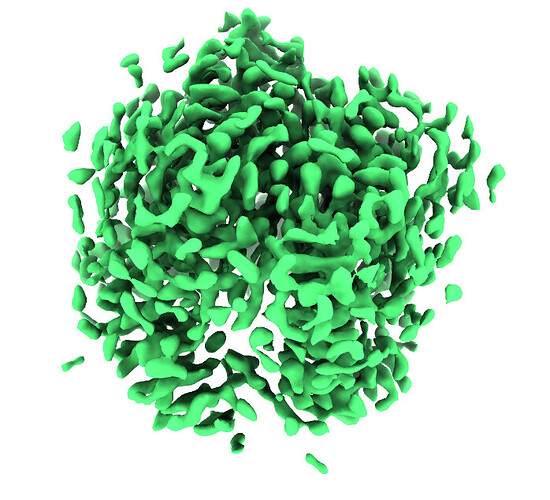
Homo Refine with ‘tv1_f01-30.mrc’
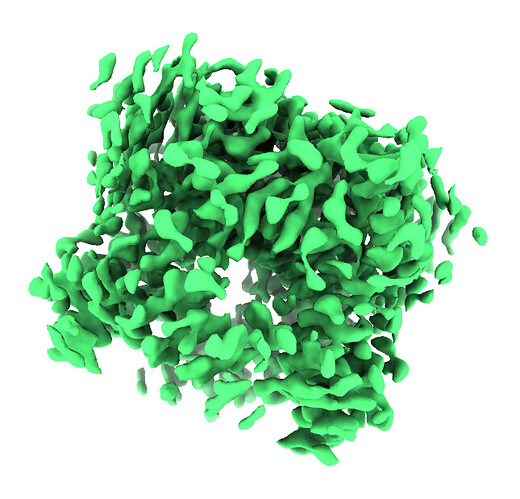
Homo Refine with ‘tv1_f03-16.mrc’
And the warning message has nothing to do with the ctf thins:
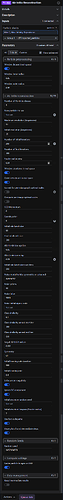
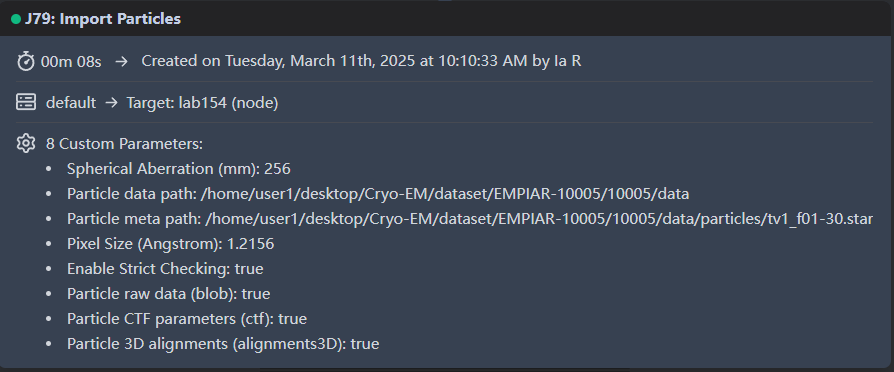
I wonder is there something wrong with the parameters I entered when importing the particles:
And may I ask which version of cryoSPARC are you using? Mine is 4.5.3, maybe different versions of cryoscan would use different methods to process data from RELION.
You’ve set the Cs (sperical aberration) to 256. There’s your issue!
Not sure what the Polara Cs is, though - the data was collected on a Polara according to the paper. Might be the same as a Titan at 2.7mm.
Anyway, rapid dips in the FSC curves like that are most often caused by pixel calibration being wrong, uncorrected beam tilt or typo’ing the microscope parameters like HT and Cs.
OMG!!!That’s the whole problem.
Thank you so much !!!
It has been bothering me for a whole week.
TF30’s nominal Cs is 2.0 mm.
EDIT: I’ve seen numbers (empirical?) that correct this to 2.26 or 2.3 mm though.
Cheers,
Yang
Grand, thanks! Never used a Polara; until we got a Krios I’d only ever been hands on with JEOL 'scopes.