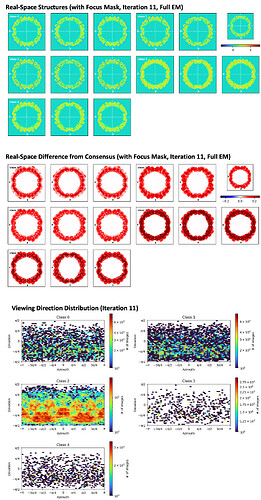
I performed 3D classification on the virus structure obtained from Homogeneous Refinement. I set the computer to divide them into 5 classes, among which Class 2 accounted for the largest proportion at 81%, and Class 3 had the deepest red color. I do not know what the varying depths of color represent, or what the viewing direction distribution the bottom picture means. Does the figure indicate that the surface proteins in Class 2 exhibit more variability?
Hello Barb, red and blue in the middle part of the figure indicate differences in density relative to the consensus, just for you to have an idea of the moving parts. In your case this is hard to tell, because the mass is so thin. The bottom part shows the number of particles in each view. Because your particles are symmetric (do I reckon the 3- and 5-fold axes from flaviviruses??? ![]() - I worked in Virology a long time ago, never did cryoEM on them), you have patches of red (=many particles contributing to the same orientation) repeting along the x axis of the plot. Class 2 has way more particles than the other classes, so maybe that is a predominant class, or the classification is not properly working for what you want to do. If you are just looking for a higher resolution reconstruction, you can try refining Class 2 alone. Classes 3 and 4 will probably give bad reconstructions (not enough particles). Not sure about 0 and 1, though. Maybe at this point you can try applying symmetry to the refinement, to see. And certainly other people in the community will give you more useful comments.
- I worked in Virology a long time ago, never did cryoEM on them), you have patches of red (=many particles contributing to the same orientation) repeting along the x axis of the plot. Class 2 has way more particles than the other classes, so maybe that is a predominant class, or the classification is not properly working for what you want to do. If you are just looking for a higher resolution reconstruction, you can try refining Class 2 alone. Classes 3 and 4 will probably give bad reconstructions (not enough particles). Not sure about 0 and 1, though. Maybe at this point you can try applying symmetry to the refinement, to see. And certainly other people in the community will give you more useful comments.
Thank you for providing the your advice, Carlos. This is the full virion structure of enterovirus. The rationale behind performing the 3D classification is to gain insights into the infectious state of the viruses. I want to know whether there are differences in the orientation of the capsid proteins among the classes, and whether certain class are more likely to infect cells than others. The underlying hypothesis is that there might be subtle structural variations in the capsid proteins between different classes, which could potentially influence their infectivity. By analysing these classes, I hope to unravel the structural determinants that govern the infectious capability of the virus particles. ![]()
You mean the envelope, or really the capsid? From the look of your classes, it seems to me the capsid will be hard to get with this approach - and I’m afraid I won’t be able to help you there.
I mean the surface protein. The enterovirus capsid is composed of four key viral proteins: VP1, VP2, VP3, and VP4. VP1, VP2, and VP3 come together to form an icosahedral shell with quasi-trimeric symmetry, while VP4 creates a network on the capsid’s interior.
Looks like ~all particles going to one class. In a highly symmetric case like this, you may want to perform symmetry expansion first followed by 3D classification with a focus mask around a single asymmetric unit - otherwise the number of classes you need to resolve variability will be very large.
Sorry, my bad. There is no envelope in Enteroviruses. Anyways, follow wathever Olib says! He’s the guy, seriously!