Hi
I am trying to locally refine a domain of my protein after NU refinement but suspect that particle subtraction is incomplete.
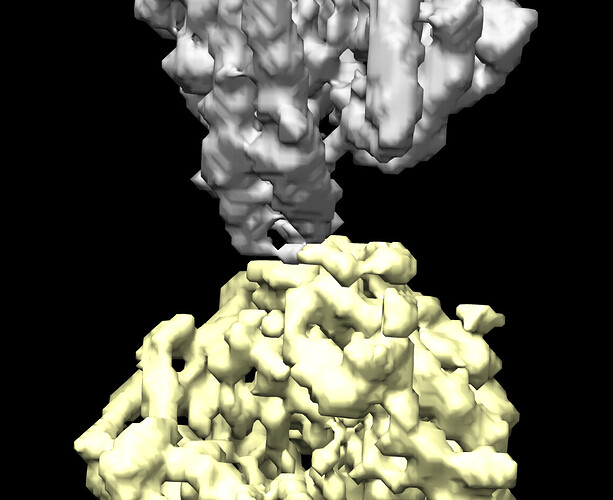
The protein has two domains and I created a mask for subtraction of one domain using Segger in Chimera with the Extract Densities from Regions tool to create .mrc files. The .mrc files for each domain at the thresholds shown below were imported to cryosparc and masks created for subtraction using these thresholds, lowpass 8A, dilation 0, soft padding 12 and fill holes enabled.
Mask created for domain 1:
Mask created for domain 2:
My settings for subtraction were:
- Input particles and volume from consensus NU refine
- Mask from above (included density for single domain)
- Window same as for NU refine
- Default settings except lowpass filter input 8A (my particle is small and difficult to see so I thought this may improve subtraction - was this correct?)
Subtraction didn’t give any error but I can’t see my particle so don’t know if it worked.
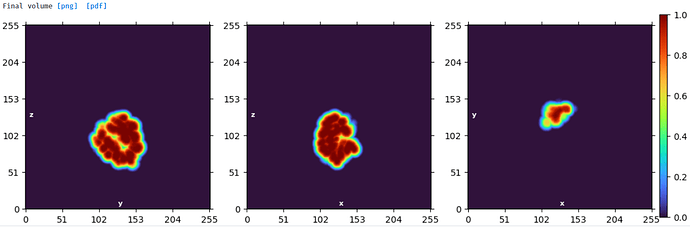
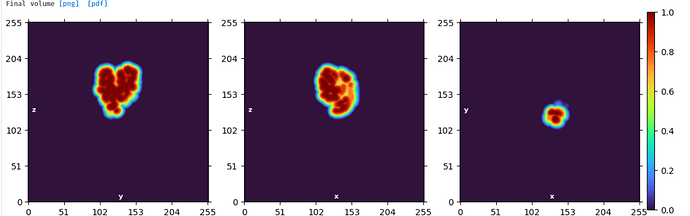
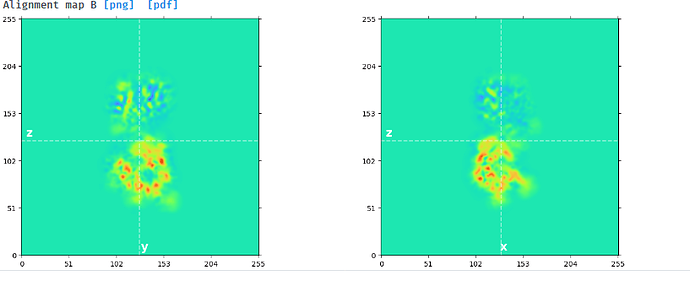
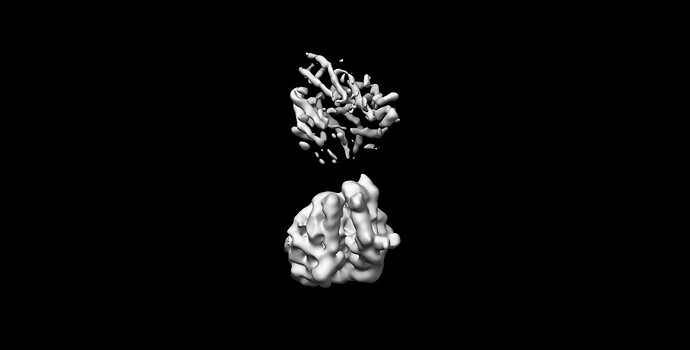
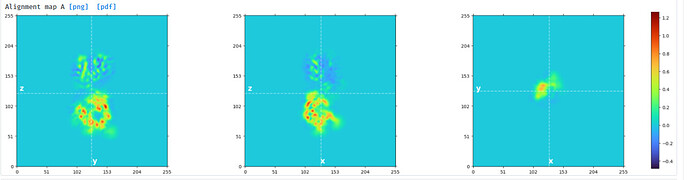
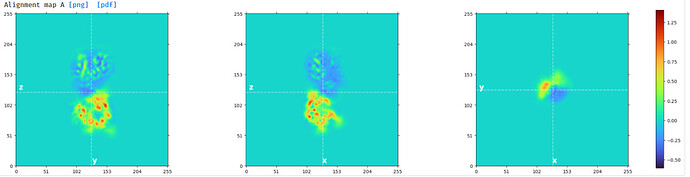
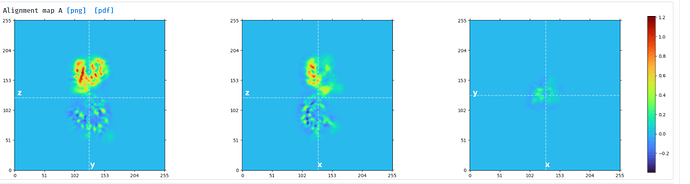
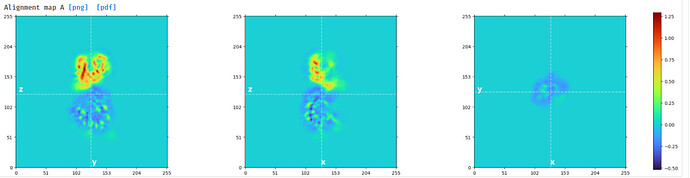
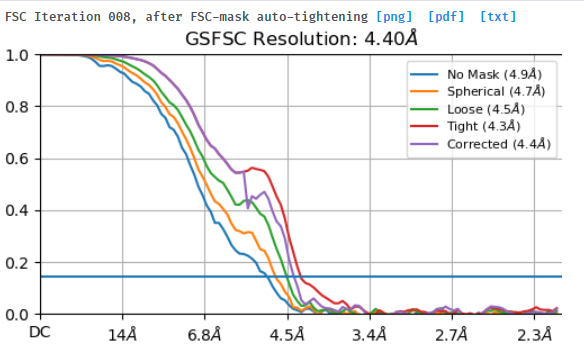
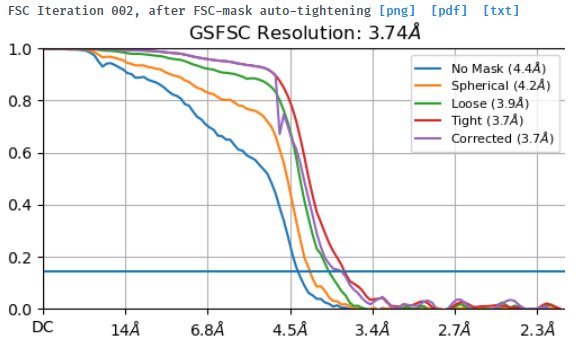
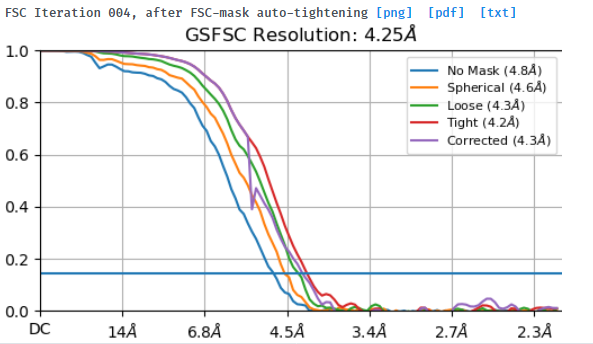
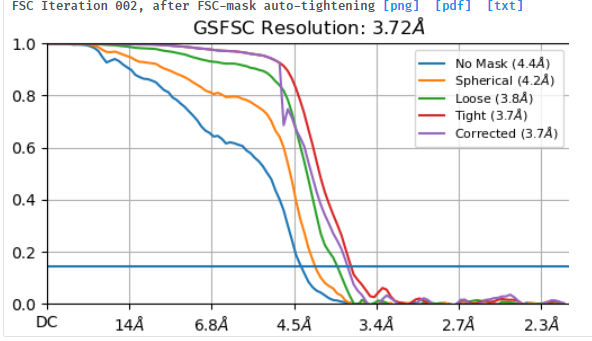
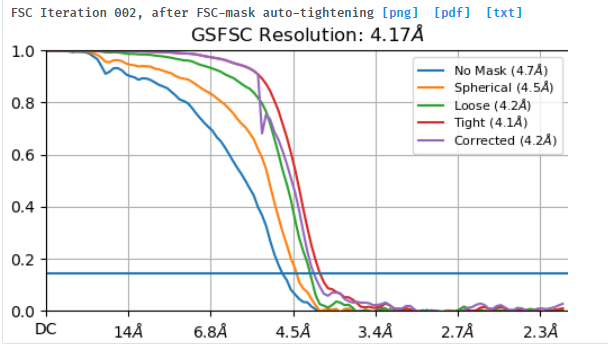
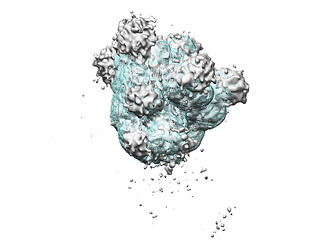
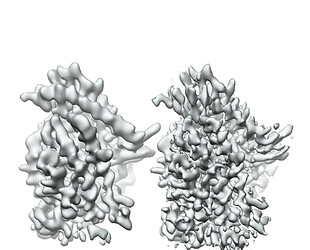
I tried to do local refine of each domain individually using a mask containing a single domain plus its glycans (created using local-filter map and Segger) with a soft edge of 12. It ended up giving artifacts at the linker between the domains. Thinking that the flexibility at this linker may have caused suboptimal subtraction of some particles, I now performed local refine using the consensus mask around the subtracted particles. It looks like the subtraction was indeed incomplete (top domain below):
Any idea how I can improve the subtraction?