Just to check one thing in that workflow, would I use the 2-domain volume from consensus NU refine as reference when doing local refine of a single domain w/o subtraction? Or should it be a single-domain reference?
I would use the two domain volume straight out of the consensus refinement as the starting point for local refinement without subtraction, but it shouldn’t make a huge difference either way
Okay. Just asked because I was using a Segger volume from Extract Densities output before and noticed today that when I lowpass it using a Volume Tools job it actually has outward ringing in real-space slices. Also visible in Chimera as a shell of density around the protein and I was wondering if that was incorrect and I should’ve used the consensus NU volume as reference for refine with subtraction? The reason why I chose the Segger one before was that I didn’t want to have a reference at 5A lowpass with noise eventually being fitted to the reference where the domain was subtracted.
I would use a direct reconstruction from the subtracted particles as your initial model for refining after subtraction - that should be the best initial model I would think, with minimal extra density in the region you have subtracted.
I wouldn’t typically use a volume treated like you describe as an initial model - particularly not when you have to use such a minimal lowpass to initialize local refinement. I would worry that any (presumably binary or at least fairly harsh) masking that Segger is doing might cause problems during refinement.
Using a volume generated by Homogeneous Reconstruction from the subtracted particles should be a straightforward way to address that problem.
Cheers
Oli
It definitely looked like Segger was doing something strange today even when I added a soft fall-off to the volume. I will stick to the reconstruction output and use that as initial reference. If the other domain is blurred it probably won’t have much of an effect.
You can use the refinement output, or just use a reconstruction from the subtracted particles, which has the advantage that the other domain will be already subtracted in the initial model.
Ah yes I get it. The consensus refinement as ref for local w/o subtraction to focus the subtraction, then homo reconstruct after subtraction and use that output as ref for local. Sorry it is midnight here and it’s been a long day so I wasn’t thinking clearly 
yep, you got it! 
HI @olibclarke,
I followed your advice for the particle subtraction and it was a really good idea 
The local refine w/o subtraction gave good alignment of the domain to be subtracted and although CS still did not remove all the density, it was much better than before.
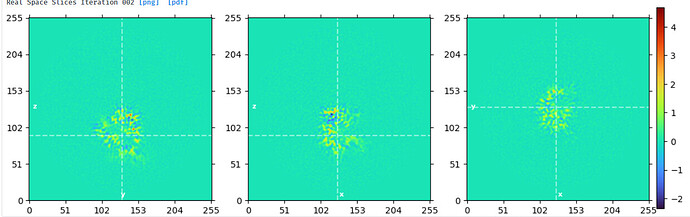
Align of bottom domain w/o subtract:
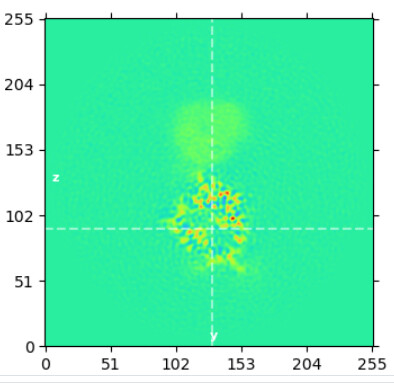
align of top domain w/o subtract:
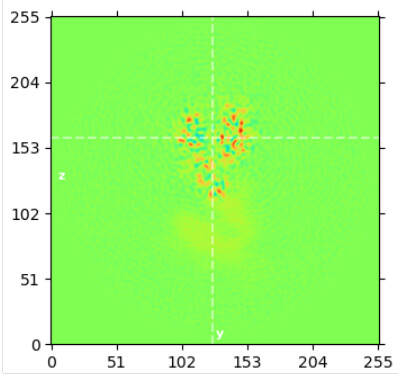
The ptcls, volume and mask from this local refine of the top domain was used for subtraction using default settings in CS.
Reconstruct using subtracted ptcls with align3D from consensus NU refine and mask around bottom domain (a bit of top domain remained):
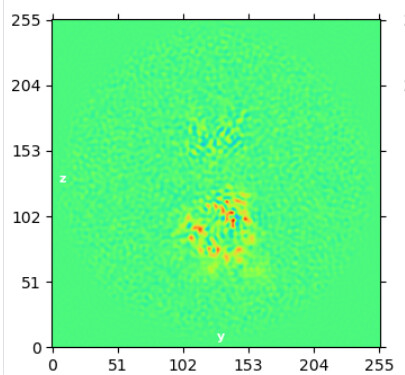
I ran local refine on these ptcls using the reconstruct volume as ini model but got overfitting issues. I have tried to optimize the mask, lowpass, priors, and search range. It seems like I need a wide mask to avoid shells of density from forming around each glycan but this wide mask seems to cause overfitting to noise - maybe because the wide mask included so much solvent?
Local refine with a lowpass 10, dilate 5, pad 16 mask which covered the glycans:
To me, the streaks in these slices looked like overfitting to noise.
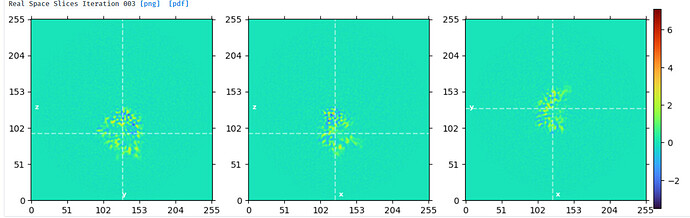
I therefore decided to run subtraction on the subtracted particles (after CS local refine) in Relion. I ran one iteration of Class3D w/o align to create an optimiser.star file to use in Relion subtraction (GUI) then ran particle subtraction with a mask around the domain to keep. I created it so that the glycans are mostly excluded but used a softly padded mask. Subsequent reconstruct in CS showed minimal glycan density and I could locally refine the single domain.
Reconstruct output after Relion subtraction (AC0.1):
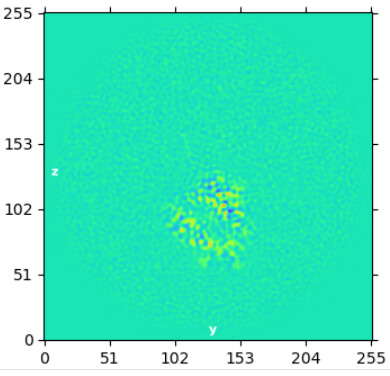
This was local without glycans (3x2 search, 9/3 priors, lowpass ini 5A:
With the protein-only refine after Relion subtraction, I had to use the alignments from localrefine where glycans were present since overwriting the align3D field after importing Relion-subtracted particles gave an error about missing fields. Subtracting glycans using the consensus refinement’s align3D didn’t seem like a good idea since this may cause incomplete glycan subtraction. So if overfitting to noise happened when the glycans were there, it could’ve been carried over. I used the reconstruct volume with initial consensus align3D as reference for protein-only refine so maybe that minimized it. This last protein-only refine looks better to my eyes - I just hope the reviewers will agree one-day 
Thanks for all the help Oli and sorry for the long-winded post!
Glad it helped! agree your last map looks improved - looks like there are some clear secondary structural features there now?
Not a suggestion re cryosparc per se, but in some cases I have found deepEMhancer (https://github.com/rsanchezgarc/deepEMhancer) useful for postprocessing in cases like this. It can be quite effective for improving map isotropy, and removing overfitting streaks. Don’t take the resultant maps as gospel though - it incorporates some kind of dust removal routine which can chew up ligands.
What is the molecular weight of each domain?
If you have plenty of particles, you may also be able to separate out some poorly aligned or subtracted particles by running class3D skipping alignments in relion (I would start with K=6, T=40), starting from your final subtracted particle stack.
Good luck!! 
Cheers
Oli
Yes the helical pitch is now visible in some areas and overall these two maps are much better than the consensus map 
They have some dust though… I haven’t played around with post-processing yet so maybe it is just from the auto sharpening. DeepEMhancer looks cool for visualization and I’ve tried it on older maps at Cosmic2 (couldn’t get it to install in Scipion) but it may be more successful with these maps. Do you usually run it with a mask? I tried without and it gave lots of noise in the box corners. Should I be concerned about that or just ignore it?
I also think that Relion Class3D may be an option. I ran it on the output from local refine of the glycoprotein after CS focused subtraction with K=3 T=20 but the classes were basically the same, just differed a bit in resolution so I kept all 100k particles together for the latest protein-only local refine. Each domain is only ~60kDa after glycan removal so not sure how low I can go i.t.o. particle numbers before SNR becomes limiting. Will give Class3D a try and maybe I’ll be pleasantly surprised!
Thank you again Oli
If you haven’t tried playing around with postprocessing much I would definitely give phenix.resolve_cryo_em a whirl too - can give very nice results on some maps! Usually I run it with the fsc_auto mask from cryosparc, like this:
phenix.resolve_cryo_em half_map_file_name_1=cryosparc_P1_J1644_002_volume_map_half_A.mrc half_map_file_name_2=cryosparc_P1_J1644_002_volume_map_half_B.mrc mask_file_name=cryosparc_P1_J1644_002_volume_mask_fsc_auto.mrc spectral_scaling=true nproc=8 soft_mask_radius=12 resolution=3.35
Deepemhancer I usually run without mask, with the highres pretrained model. Yes it gives some funkiness at low thresholds and in the corners of the map, but the improvements in the protein area can be worth the tradeoff in favourable cases.
Thanks I haven’t ever tried phenix but will give it a go!
This is a great workflow description @olibclarke – exactly the kind of thing we were hoping to make easier with the low-level results feature and with keeping the frame of reference fixed for local refinements! I’m glad to see that the process of iterating signal subtraction has been useful for this challenging dataset!
@lizellelubbe Just to help us understand the workflow (and to potentially help make this workflow easier in the future) could I confirm that you did the following:
- After following steps 1 and 2, you now had two sets of particles: set X (aligned to domain A, with domain A subtracted) and set Y (aligned to domain B, with domain B subtracted)
- Then you took set X and set Y (which have already each had one round of subtraction in cryoSPARC) and performed subtraction in RELION such that the glycans were subtracted out? E.g. so that set X has only domain B (with glycans and domain A both subtracted out)?
- Then finally, you used these new sets for final local refinements in cryoSPARC
Best,
Michael
Yes it is great! Do you have a short video or something with gifs describing the low level results interface? Once you get it it makes sense, but it is a little hard to describe without showing people.
Also the key aspect that a particle blob is formed from the intersection of the components, so you can easily use a selection of downsampled or subtracted particles to get original particles, is something that makes it very powerful and frequently used for me.
Hi @mmclean,
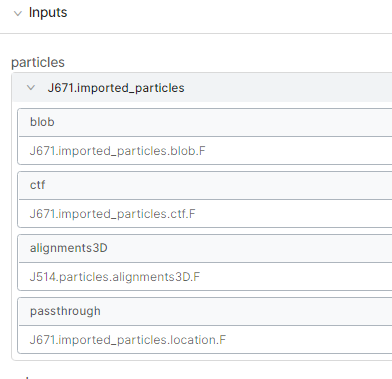
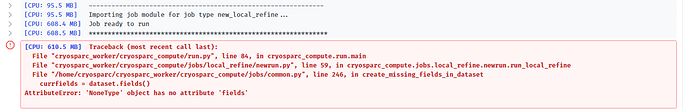
Yes, that is basically what I did. But after doing steps 1-2 and performing homogenous reconstruct to recover a volume of the non-subtracted domain with the consensus align3D, I ran local refine in CS while the glycans were still there before going into Relion. I thought that it might allow better subtraction of glycan densities if the protein alignment was improved over the consensus. I then wanted to use the consensus align3D again for local refine after glycan subtraction since the local refine with glycans gave overfitting streaks. This failed and I couldn’t figure out why. The Relion (GUI) altered my star file columns and it gave a warning in the output log that the optics groups are in the wrong order and being renamed so maybe that caused it. Other than that, the method suggested by @olibclarke worked really well!
Input to local refine after glycan subtraction:
star file input to Relion subtraction:
data_optics
loop_
_rlnVoltage #1
_rlnSphericalAberration #2
_rlnAmplitudeContrast #3
_rlnOpticsGroup #4
_rlnImageSize #5
_rlnImagePixelSize #6
_rlnImageDimensionality #7
300.000000 2.700000 0.100000 4 256 1.060000 2
data_particles
loop_
_rlnImageName #1
_rlnMicrographName #2
_rlnCoordinateX #3
_rlnCoordinateY #4
_rlnAngleRot #5
_rlnAngleTilt #6
_rlnAnglePsi #7
_rlnOriginXAngst #8
_rlnOriginYAngst #9
_rlnDefocusU #10
_rlnDefocusV #11
_rlnDefocusAngle #12
_rlnPhaseShift #13
_rlnCtfBfactor #14
_rlnOpticsGroup #15
_rlnRandomSubset #16
_rlnClassNumber #17
000001@J630/subtracted_particles_batch_0A.mrcs J10/imported/FoilHole_8065276_Data_7120079_7120081_20191008_0757_fractions.mrc 3660 1832 -173.511185 58.548630 -23.863300 1.384657 3.299146 18634.656250 18421.523438 245.198395 0.000000 0.000000 4 1 1
star file after Relion subtraction:
version 30001
data_optics
loop_
_rlnVoltage #1
_rlnSphericalAberration #2
_rlnAmplitudeContrast #3
_rlnOpticsGroup #4
_rlnImageSize #5
_rlnImagePixelSize #6
_rlnImageDimensionality #7
300.000000 2.700000 0.100000 1 256 1.060000 2
version 30001
data_particles
loop_
_rlnImageName #1
_rlnMicrographName #2
_rlnCoordinateX #3
_rlnCoordinateY #4
_rlnAngleRot #5
_rlnAngleTilt #6
_rlnAnglePsi #7
_rlnOriginXAngst #8
_rlnOriginYAngst #9
_rlnDefocusU #10
_rlnDefocusV #11
_rlnDefocusAngle #12
_rlnPhaseShift #13
_rlnCtfBfactor #14
_rlnOpticsGroup #15
_rlnRandomSubset #16
_rlnClassNumber #17
_rlnGroupNumber #18
_rlnNormCorrection #19
_rlnLogLikeliContribution #20
_rlnMaxValueProbDistribution #21
_rlnNrOfSignificantSamples #22
_rlnImageOriginalName #23
000001@Subtract/job931/Particles/subtracted_rank1_opticsgroup1.mrcs J10/imported/FoilHole_10146744_Data_7120079_7120081_20191010_0954_fractions.mrc 3379.000000 4481.000000 -65.48501 78.080360 -117.26398 3.898997 -2.00063 12901.792969 12812.845703 -45.42252 0.000000 0.000000 1 1 1 1 0.918014 0.083082 1.000000 1 000424@J630/subtracted_particles_batch_25A.mrcs
error when using consensus align3D after Relion subtraction:
Dear Oli,
Thanks for this very great idea. For the 3, instead of using the orientations from NU refine, how about using the orientations from the local refinement on domain B from 1?
Thanks and best
Yep no reason why not
thank you so much again